kfoldLoss
交差検証された分割済みの回帰モデルの損失
説明
例
carsmall データのアンサンブル回帰の交差検証損失を求めます。
carsmall データ セットを読み込み、排気量、馬力および車両重量を予測子として選択します。
load carsmall
X = [Displacement Horsepower Weight];回帰木のアンサンブルに学習をさせます。
rens = fitrensemble(X,MPG);
rens から交差検証済みのアンサンブルを作成し、k 分割交差検証損失を求めます。
rng(10,'twister') % For reproducibility cvrens = crossval(rens); L = kfoldLoss(cvrens)
L = 28.7114
平均二乗誤差 (MSE) はモデルの品質を示す尺度です。交差検証済み回帰モデルの各分割の MSE を調べます。
carsmall データ セットを読み込みます。予測子 X と応答データ Y を指定します。
load carsmall
X = [Cylinders Displacement Horsepower Weight];
Y = MPG;交差検証済みの回帰木モデルの学習を行います。既定では、10 分割交差検証が実行されます。
rng('default') % For reproducibility CVMdl = fitrtree(X,Y,'CrossVal','on');
各分割の MSE を計算します。箱ひげ図を使用して損失値の分布を可視化します。いずれの値も外れ値でないことがわかります。
losses = kfoldLoss(CVMdl,'Mode','individual')
losses = 10×1
42.5072
20.3995
22.3737
34.4255
40.8005
60.2755
19.5562
9.2060
29.0788
16.3386
boxchart(losses)

交差検証済みの 10 分割の一般化加法モデル (GAM) に学習させます。その後、kfoldLoss を使用して交差検証の累積回帰損失 (平均二乗誤差) を計算します。誤差を使用して、予測子 (予測子の線形項) あたりの最適な木の数と交互作用項あたりの最適な木の数を特定します。
代わりに、名前と値の引数OptimizeHyperparametersを使用して fitrgam の名前と値の引数の最適な値を特定することもできます。例については、OptimizeHyperparameters を使用した GAM の最適化を参照してください。
patients データ セットを読み込みます。
load patients予測子変数 (Age、Diastolic、Smoker、Weight、Gender、および SelfAssessedHealthStatus) と応答変数 (Systolic) を格納する table を作成します。
tbl = table(Age,Diastolic,Smoker,Weight,Gender,SelfAssessedHealthStatus,Systolic);
既定の交差検証オプションを使用して交差検証済み GAM を作成します。名前と値の引数 'CrossVal' を 'on' として指定します。また、5 つの交互作用項を含めるように指定します。
rng('default') % For reproducibility CVMdl = fitrgam(tbl,'Systolic','CrossVal','on','Interactions',5);
'Mode' を 'cumulative' として指定すると、関数 kfoldLoss は累積誤差を返します。これは、各分割に同じ数の木を使用して取得したすべての分割の平均誤差です。各分割の木の数を表示します。
CVMdl.NumTrainedPerFold
ans = struct with fields:
PredictorTrees: [300 300 300 300 300 300 300 300 300 300]
InteractionTrees: [76 100 100 100 100 42 100 100 59 100]
kfoldLoss では、最大で 300 個の予測子木と 42 個の交互作用木を使用して累積誤差を計算できます。
10 分割交差検証を行った累積平均二乗誤差をプロットします。'IncludeInteractions' を false として指定して、計算から交互作用項を除外します。
L_noInteractions = kfoldLoss(CVMdl,'Mode','cumulative','IncludeInteractions',false); figure plot(0:min(CVMdl.NumTrainedPerFold.PredictorTrees),L_noInteractions)
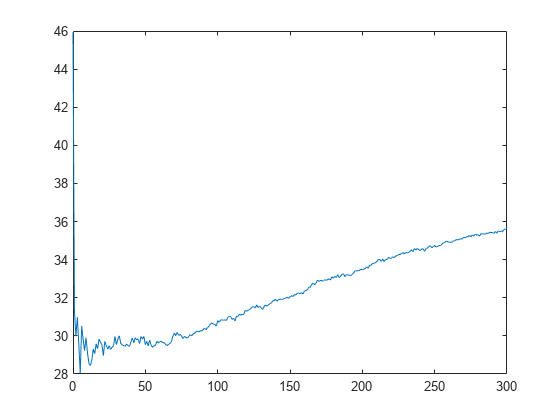
L_noInteractions の最初の要素は、切片 (定数) 項のみを使用して取得したすべての分割の平均誤差です。L_noInteractions の (J+1) 番目の要素は、切片項と各線形項の最初の J 個の予測子木を使用して取得した平均誤差です。累積損失をプロットすると、GAM の予測子木の数が増えるにつれて誤差がどのように変化するかを観察できます。
最小誤差とその最小誤差の達成時に使用された予測子木の数を調べます。
[M,I] = min(L_noInteractions)
M = 28.0506
I = 6
GAM に 5 個の予測子木が含まれるときに誤差が最小になっています。
線形項と交互作用項の両方を使用して累積平均二乗誤差を計算します。
L = kfoldLoss(CVMdl,'Mode','cumulative'); figure plot(0:min(CVMdl.NumTrainedPerFold.InteractionTrees),L)
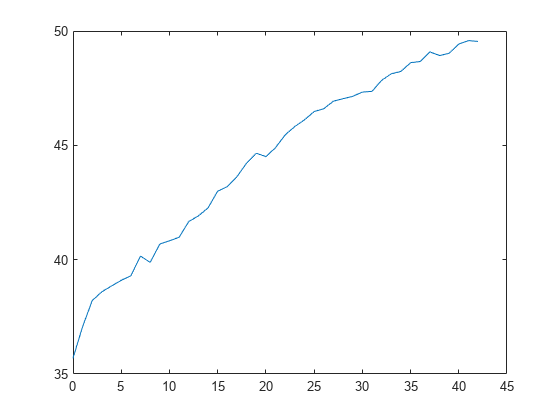
L の最初の要素は、切片 (定数) 項と各線形項のすべての予測子木を使用して取得したすべての分割の平均誤差です。L の (J+1) 番目の要素は、切片項、各線形項のすべての予測子木、および各交互作用項の最初の J 個の交互作用木を使用して取得した平均誤差です。プロットから、交互作用項を追加すると誤差が大きくなることがわかります。
予測子木の数が 5 個のときの誤差で問題がなければ、一変量の GAM にもう一度学習させ、交差検証を使用せずに 'NumTreesPerPredictor',5 と指定して予測モデルを作成できます。
入力引数
交差検証された分割済みの回帰モデル。RegressionPartitionedModel オブジェクト、RegressionPartitionedEnsemble オブジェクト、RegressionPartitionedGAM オブジェクト、RegressionPartitionedGP オブジェクト、RegressionPartitionedNeuralNetwork オブジェクト、または RegressionPartitionedSVM オブジェクトとして指定します。オブジェクトは 2 つの方法で作成できます。
次の表に記載されている学習済み回帰モデルをそのオブジェクト関数
crossvalに渡す。次の表に記載されている関数を使用して回帰モデルに学習をさせ、その関数の交差検証に関する名前と値の引数のいずれかを指定する。
| 回帰モデル | 関数 |
|---|---|
RegressionEnsemble | fitrensemble |
RegressionGAM | fitrgam |
RegressionGP | fitrgp |
RegressionNeuralNetwork | fitrnet |
RegressionSVM | fitrsvm |
RegressionTree | fitrtree |
名前と値の引数
オプションの引数のペアを Name1=Value1,...,NameN=ValueN として指定します。ここで、Name は引数名で、Value は対応する値です。名前と値の引数は他の引数の後に指定しなければなりませんが、ペアの順序は重要ではありません。
R2021a より前では、名前と値をそれぞれコンマを使って区切り、Name を引用符で囲みます。
例: kfoldLoss(CVMdl,'Folds',[1 2 3 5]) は、平均二乗誤差の計算に 1 番目、2 番目、3 番目、および 5 番目の分割を使用し、4 番目の分割は除外するように指定します。
使用する分割のインデックス。正の整数ベクトルとして指定します。Folds の要素は 1 から CVMdl.KFold の範囲でなければなりません。
Folds で指定された分割のみが使用されます。
例: 'Folds',[1 4 10]
データ型: single | double
モデルの交互作用項を含むというフラグ。true または false として指定します。この引数は、一般化加法モデル (GAM) の場合のみ有効です。つまり、この引数を指定できるのは、CVMdl が RegressionPartitionedGAM である場合だけです。
CVMdl のモデル (CVMdl.Trained) に交互作用項が含まれる場合、既定値は true です。モデルに交互作用項が含まれない場合、値は false でなければなりません。
例: 'IncludeInteractions',false
データ型: logical
損失関数。'mse' または関数ハンドルとして指定します。
組み込み関数
'mse'を指定します。この場合、損失関数は平均二乗誤差です。関数ハンドル表記を使用して独自の関数を指定します。
n は学習データの観測値数 (
CVMdl.NumObservations) とします。使用する関数ではシグネチャがlossvalue =になっていなければなりません。ここでlossfun(Y,Yfit,W)出力引数
lossvalueはスカラーです。関数名 (
lossfun) を指定します。Yは、観測された応答の n 行 1 列の数値ベクトルです。Yfitは、予測された応答の n 行 1 列の数値ベクトルです。Wは、観測値の重みの n 行 1 列の数値ベクトルです。
'LossFun',@を使用して独自の関数を指定します。lossfun
データ型: char | string | function_handle
出力の集約レベル。'average'、'individual'、または 'cumulative' として指定します。
| 値 | 説明 |
|---|---|
'average' | 出力は、すべての分割の平均を表すスカラー値です。 |
'individual' | 出力は、分割ごとに 1 つずつの値が含まれている長さ k のベクトルです。k は分割数です。 |
'cumulative' | メモ この値を指定する場合、
|
例: 'Mode','individual'
R2023b 以降
予測子に欠損値がある観測値に使用する予測した応答値。"median"、"mean"、"omitted"、または数値スカラーとして指定します。この引数は、ガウス過程回帰モデル、ニューラル ネットワーク モデル、サポート ベクター マシン モデルに対してのみ有効です。つまり、この引数を指定できるのは、CVMdl が RegressionPartitionedGP オブジェクト、RegressionPartitionedNeuralNetwork オブジェクト、または RegressionPartitionedSVM オブジェクトである場合だけです。
| 値 | 説明 |
|---|---|
"median" |
この値は、 |
"mean" | kfoldLoss は、予測子に欠損値がある観測値について予測した応答値として、学習分割データ内の観測された応答値の平均値を使用します。 |
"omitted" | kfoldLoss は、予測子に欠損値がある観測値を損失の計算から除外します。 |
| 数値スカラー | kfoldLoss は、予測子に欠損値がある観測値について予測した応答値として、この値を使用します。 |
観測された応答値または観測値の重みが観測値にない場合、その観測値は kfoldLoss による損失の計算に使用されません。
例: "PredictionForMissingValue","omitted"
データ型: single | double | char | string
出力引数
損失。数値スカラーまたは数値列ベクトルとして返されます。
既定の損失は、検証分割観測値と、学習分割観測値で学習させた回帰モデルによる予測との間の平均二乗誤差です。
Modeが'average'である場合、Lはすべての分割の平均損失です。Modeが'individual'である場合、Lは各分割の損失が含まれている k 行 1 列の数値列ベクトルです。k は分割数です。Modeが'cumulative'でCVMdlがRegressionPartitionedEnsembleである場合、Lはmin(CVMdl.NumTrainedPerFold)行 1 列の数値列ベクトルです。各要素jは、弱学習器1:jで学習させたアンサンブルを使用して取得したすべての分割の平均損失です。Modeが'cumulative'でCVMdlがRegressionPartitionedGAMの場合、出力の値はIncludeInteractionsの値によって異なります。IncludeInteractionsがfalseの場合、Lは(1 + min(NumTrainedPerFold.PredictorTrees))行 1 列の数値列ベクトルです。Lの最初の要素は、切片 (定数) 項のみを使用して取得したすべての分割の平均損失です。Lの(j + 1)番目の要素は、切片項と各線形項の最初のj個の予測子木を使用して取得した平均損失です。IncludeInteractionsがtrueの場合、Lは(1 + min(NumTrainedPerFold.InteractionTrees))行 1 列の数値列ベクトルです。Lの最初の要素は、切片 (定数) 項と各線形項のすべての予測子木を使用して取得したすべての分割の平均損失です。Lの(j + 1)番目の要素は、切片項、各線形項のすべての予測子木、および各交互作用項の最初のj個の交互作用木を使用して取得した平均損失です。
代替機能
木モデルの交差検証損失を計算する場合は、cvloss を呼び出すことで RegressionPartitionedModel オブジェクトの作成を回避できます。調査を複数回行う予定の場合、交差検証木オブジェクトを作成して時間を節約できます。
拡張機能
使用上の注意および制限:
この関数は、次のモデルの GPU 配列を完全にサポートします。
fitrtreeを使用するかRegressionTreeオブジェクトをcrossvalに渡すことによって当てはめたRegressionPartitionedModelオブジェクト
詳細は、GPU での MATLAB 関数の実行 (Parallel Computing Toolbox)を参照してください。
バージョン履歴
R2011a で導入kfoldLoss で RegressionPartitionedNeuralNetwork モデルの GPU 配列が完全にサポートされます。
R2023b 以降で損失を予測または計算する際、一部の回帰モデルでは、予測子に欠損値がある観測値について予測した応答値を指定できます。名前と値の引数 PredictionForMissingValue を指定して、予測値として数値スカラー、学習セットの中央値、または学習セットの平均値を使用します。損失を計算するときに、予測子に欠損値がある観測値を省略するように指定することもできます。
次の表は、名前と値の引数 PredictionForMissingValue をサポートするオブジェクト関数の一覧です。既定では、これらの関数は、予測子に欠損値がある観測値について予測した応答値として、学習セットの中央値を使用します。
| モデル タイプ | モデル オブジェクト | オブジェクト関数 |
|---|---|---|
| ガウス過程回帰 (GPR) モデル | RegressionGP, CompactRegressionGP | loss, predict, resubLoss, resubPredict |
RegressionPartitionedGP | kfoldLoss, kfoldPredict | |
| ガウス カーネル回帰モデル | RegressionKernel | loss, predict |
RegressionPartitionedKernel | kfoldLoss, kfoldPredict | |
| 線形回帰モデル | RegressionLinear | loss, predict |
RegressionPartitionedLinear | kfoldLoss, kfoldPredict | |
| ニューラル ネットワーク回帰モデル | RegressionNeuralNetwork, CompactRegressionNeuralNetwork | loss, predict, resubLoss, resubPredict |
RegressionPartitionedNeuralNetwork | kfoldLoss, kfoldPredict | |
| サポート ベクター マシン (SVM) 回帰モデル | RegressionSVM, CompactRegressionSVM | loss, predict, resubLoss, resubPredict |
RegressionPartitionedSVM | kfoldLoss, kfoldPredict |
以前のリリースでは、上記の回帰モデル関数 loss および predict は、予測子に欠損値がある観測値について予測した応答値として NaN を使用していました。予測子に欠損値がある観測値は、予測と損失の再代入 ("resub") と交差検証 ("kfold") の計算で省略されていました。
R2023a 以降では、kfoldLoss で RegressionPartitionedSVM モデルの GPU 配列が完全にサポートされます。
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Web サイトの選択
Web サイトを選択すると、翻訳されたコンテンツにアクセスし、地域のイベントやサービスを確認できます。現在の位置情報に基づき、次のサイトの選択を推奨します:
また、以下のリストから Web サイトを選択することもできます。
最適なサイトパフォーマンスの取得方法
中国のサイト (中国語または英語) を選択することで、最適なサイトパフォーマンスが得られます。その他の国の MathWorks のサイトは、お客様の地域からのアクセスが最適化されていません。
南北アメリカ
- América Latina (Español)
- Canada (English)
- United States (English)
ヨーロッパ
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)