spectralMatch
スペクトル ライブラリを使用した未知の領域または物質の識別
構文
説明
score = spectralMatch(libData,reflectance,wavelength)reflectance および wavelength として指定された分光反射率値を ECOSTRESS スペクトル ライブラリ libData で入手できる値と照合し、領域または物質を識別します。
score = spectralMatch(___,Name=Value)
メモ
この関数には、Hyperspectral Imaging Library for Image Processing Toolbox™ が必要です。Hyperspectral Imaging Library for Image Processing Toolbox はアドオン エクスプローラーからインストールできます。アドオンのインストールの詳細については、アドオンの入手と管理を参照してください。
Hyperspectral Imaging Library for Image Processing Toolbox は、MATLAB® Online™ および MATLAB Mobile™ によってサポートされないため、デスクトップの MATLAB が必要です。
例
スペクトル マッチング法では、ハイパースペクトル データ キューブ内にある各ピクセルのスペクトル シグネチャを、ECOSTRESS スペクトル ファイルの植生の基準スペクトル シグネチャと比較します。
ECOSTRESS スペクトル ライブラリから植生のスペクトル シグネチャを読み取ります。
filename = "vegetation.tree.tsuga.canadensis.vswir.tsca-1-47.ucsb.asd.spectrum.txt";
libData = readEcostressSig(filename);ハイパースペクトル データをワークスペースに読み取ります。
hcube = imhypercube("paviaU.hdr");基準スペクトルに対する、ハイパースペクトル データ ピクセルのスペクトルの距離スコアを計算します。
score = spectralMatch(libData,hcube);
距離スコアを表示します。距離スコアの小さいピクセルは基準スペクトルと強く一致するため、植生領域に属している可能性が高くなります。
imagesc(score) colorbar
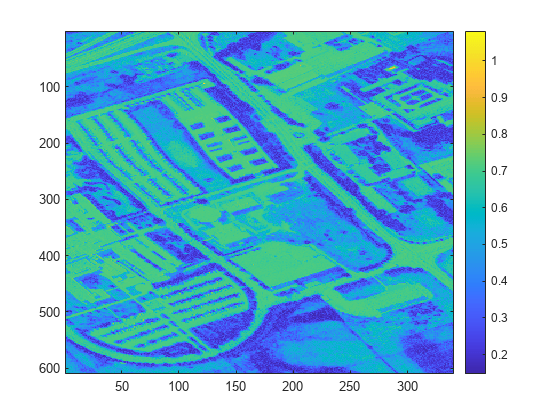
植生領域に対応する距離スコアを検出するためのしきい値を定義します。
threshold = 0.3;
しきい値を適用してバイナリ イメージを作成します。値が 1 のバイナリ イメージ内の領域は、距離スコアがしきい値未満のデータ キューブ内の植生領域に対応します。その他のすべてのピクセルは値が 0 になります。
bw = score < threshold;
バイナリ イメージ内の最大強度の領域のインデックスを使用して、ハイパースペクトル データ キューブの植生領域をセグメント化します。
datacube = gather(hcube); T = reshape(datacube,[size(datacube,1)*size(datacube,2) size(datacube,3)]); Ts = zeros(size(T)); Ts(bw == 1,:) = T( bw==1 ,:); Ts = reshape(Ts,[size(datacube,1) size(datacube,2) size(datacube,3)]);
セグメント化された植生領域のみを含む新しい hypercube オブジェクトを作成します。
segmentedDataCube = imhypercube(Ts,hcube.Wavelength);
colorize 関数を使用して、元のデータ キューブとセグメント化されたデータ キューブの RGB カラー イメージを推定します。
rgbImg = colorize(hcube,Method="rgb",ContrastStretching=true); segmentedImg = colorize(segmentedDataCube,Method="rgb",ContrastStretching=true);
関数 imoverlay を使用して、元のデータ キューブの RGB バージョンにバイナリ イメージを重ね合わせます。
B = imoverlay(rgbImg,bw,"Yellow");元のデータ キューブとセグメント化されたデータ キューブの RGB カラー イメージ、および重ね合わせたイメージを表示します。セグメント化されたイメージには、元のデータ キューブからセグメント化された植生領域のみが含まれています。
figure
montage({rgbImg,segmentedImg,B},Size=[1 3])
title("Original Image | Segmented Image | Overlaid Image")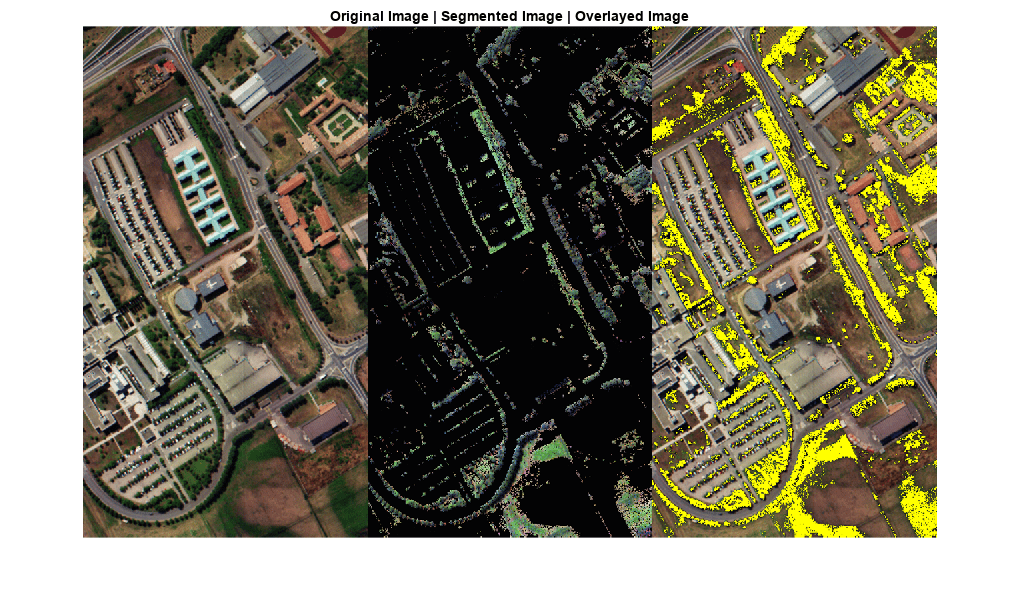
ECOSTRESS スペクトル ライブラリから基準スペクトル シグネチャを読み取ります。このライブラリは、人工物質、土壌、水、植生に属する 15 のスペクトル シグネチャで構成されています。出力は、ECOSTRESS ライブラリ ファイルから読み取ったスペクトル データを格納する構造体配列です。
dirname = fullfile(matlabroot,"toolbox","images","supportpackages","hyperspectral","hyperdata","ECOSTRESSSpectraFiles"); libData = readEcostressSig(dirname);
未知の物質の反射率と波長値を含む .mat ファイルをワークスペースに読み込みます。反射率値と波長値を合わせてテスト スペクトルが構成されます。
load spectralData.mat reflectance wavelength
スペクトル情報発散 (SID) 法を使用して、基準スペクトルとテスト スペクトルの間のスペクトルの一致を計算します。関数は、帯域幅がテスト スペクトルと重なり合う基準スペクトルのみの距離スコアを計算します。その他のスペクトルにはすべて警告メッセージが表示されます。警告メッセージはオフにできます。
warning off score = spectralMatch(libData,reflectance,wavelength,Method="SID");
テスト スペクトルの距離スコアを表示します。距離スコアの小さいピクセルは基準スペクトルと強く一致します。距離スコア値 NaN は、対応する基準スペクトルとテスト スペクトルが、帯域幅の重複部分のしきい値を満たしていないことを示します。
score
score = 1×15
297.8016 122.5567 203.5864 103.3351 288.7747 275.5321 294.2341 NaN NaN 290.4887 NaN 299.5762 171.6919 46.2072 176.6637
最小の距離スコアと対応するインデックスを求めます。返されたインデックス値は、テスト スペクトルに最も厳密に一致する基準スペクトルを含む構造体配列 libData の行を示します。
[value,ind] = min(score);
最小距離スコアのインデックスを使用して一致する基準スペクトルを求め、ECOSTRESS ライブラリ内の一致するスペクトル データの詳細を表示します。結果から、テスト スペクトルが海水のスペクトル シグネチャに最も厳密に一致することが分かります。
matchingSpectra = libData(ind)
matchingSpectra = struct with fields:
Name: "Sea Foam"
Type: "Water"
Class: "Sea Water"
SubClass: "none"
ParticleSize: "Liquid"
Genus: [0×0 string]
Species: [0×0 string]
SampleNo: "seafoam"
Owner: "Dept. of Earth and Planetary Science, John Hopkins University"
WavelengthRange: "TIR"
Origin: "JHU IR Spectroscopy Lab."
CollectionDate: "N/A"
Description: "Sea foam water. Original filename FOAM Original ASTER Spectral Library name was jhu.becknic.water.sea.none.liquid.seafoam.spectrum.txt"
Measurement: "Directional (10 Degree) Hemispherical Reflectance"
FirstColumn: "X"
SecondColumn: "Y"
WavelengthUnit: "micrometer"
DataUnit: "Reflectance (percent)"
FirstXValue: "14.0112"
LastXValue: "2.0795"
NumberOfXValues: "2110"
AdditionalInformation: "none"
Wavelength: [2110×1 double]
Reflectance: [2110×1 double]
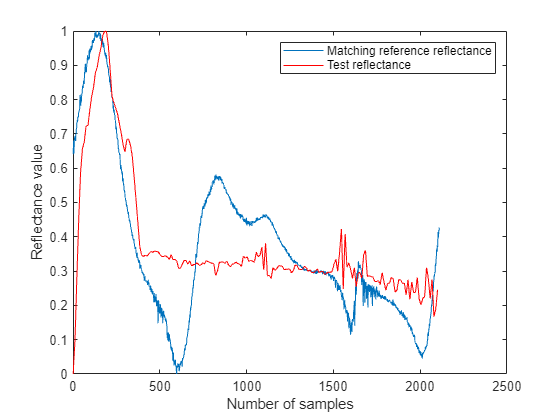
テスト スペクトルの反射率値および対応する基準スペクトルの反射率値をプロットします。反射率曲線の形状をプロットして可視化するため、反射率の値を範囲 [0, 1] に再スケーリングし、基準反射率値の合計に一致するようにテスト反射率値を内挿します。
figure testReflectance = rescale(reflectance,0,1); refReflectance = rescale(matchingSpectra.Reflectance,0,1); testLength = length(testReflectance); newLength = length(testReflectance)/length(refReflectance); testReflectance = interp1(1:testLength,testReflectance,1:newLength:testLength); plot(refReflectance) hold on plot(testReflectance,"r") hold off legend("Matching reference reflectance","Test reflectance") xlabel("Number of samples") ylabel("Reflectance value")
入力引数
スペクトル ライブラリからのスペクトル データ。構造体または 1 行 K 列の構造体配列として指定します。K は、スペクトル マッチングに使用するライブラリからのスペクトル シグネチャの数です。各スペクトル ライブラリ構造体には、関数 readEcostressSig を使用して ECOSTRESS ライブラリから読み取られたスペクトル シグネチャなど、等しい長さの数値ベクトルで構成される Reflectance フィールドと Wavelength フィールドが含まれていなければなりません。
入力ハイパースペクトル データ。hypercube オブジェクトとして指定します。
反射率値。C 要素ベクトルとして指定します。C は反射率値を測定した波長の数です。
波長値。C 要素ベクトルとして指定します。C は反射率値を測定した波長の数です。
名前と値の引数
オプションの引数のペアを Name1=Value1,...,NameN=ValueN として指定します。ここで、Name は引数名で、Value は対応する値です。名前と値の引数は他の引数の後に指定しなければなりませんが、ペアの順序は重要ではありません。
R2021a より前では、コンマを使用して名前と値をそれぞれ区切り、Name を引用符で囲みます。
例: spectralMatch(libData,hcube,MinBandWidth=0.5)
スペクトル マッチング方法。次のいずれかの値として指定します。
"sam"— スペクトル角マッパー (SAM) 法。2 つのスペクトル間の角距離を計算して、それらの類似度を評価します。"sid"— スペクトル情報発散 (SID) 法。2 つのスペクトルの確率分布値の差を計算して、それらの類似度を評価します。"sidsam"— 複合型のスペクトル類似度評価法。SID と SAM の距離尺度を組み合わせて 2 つのスペクトル間の類似度を評価します。"jmsam"— ジェフリー・松下スペクトル角マッパー (JMSAM)。ジェフリー・松下 (JM) と SAM の距離尺度を組み合わせて 2 つのスペクトル間の類似度を評価します。"ns3"— 正規化スペクトル類似度スコア (Normalized spectral similarity score、NS3) 法。ユークリッドと SAM の距離尺度を組み合わせて 2 つのスペクトル間の類似度を評価します。
これらのスペクトル マッチング法の詳細については、アルゴリズムを参照してください。
データ型: char | string
帯域幅の重複部分の最小値。ナノメートル単位の正のスカラーとして指定します。基準スペクトルとテスト スペクトルの帯域幅の重複部分は次のように定義されます。
BWoverlap = Wmax − Wmin
Wmin は、基準スペクトルおよびテスト スペクトルの最小波長の最大値です。
Wmax は、基準スペクトルおよびテスト スペクトルの最大波長の最大値です。
引数 MinBandWidth は、テスト物質のスペクトル値と ECOSTRESS スペクトル データの間の帯域幅の重複部分の最小期待値を定義します。
データ型: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
出力引数
距離スコア。3 次元数値配列、行列、K 要素の列ベクトル、またはスカラーとして返されます。出力スコアの次元は、libData の次元、およびテスト データが hypercube オブジェクトであるか wavelength と reflectance のペアであるかによって異なります。
テスト スペクトル シグネチャが hypercube オブジェクトとして指定された場合、hcube およびデータ キューブのサイズは M x N x C となります。
入力引数の次元 (libData) | 出力の次元 (score) |
| 1 行 K 列。K 個のスペクトル ファイルから読み取った K 個の基準シグネチャを含む | サイズが M x N x K の 3 次元数値配列。K 個の基準シグネチャを基準とするピクセルごとの距離スコアを含む K の各チャネルの値は、 |
| 1 行 1 列。1 つのスペクトル ファイル (K = 1) から読み取った基準シグネチャを含む | サイズが M 行 N 列の行列。基準シグネチャを基準とするピクセルごとのスペクトルの距離スコアを含む。 |
テスト スペクトル シグネチャが reflectance および wavelength の値で指定された場合は、次のようになります。
入力引数の次元 (libData) | 出力の次元 (score) |
| 1 行 K 列。K 個のスペクトル ファイルから読み取った K 個の基準シグネチャを含む | K 要素のベクトル。K 個の基準シグネチャを基準とするテスト スペクトルの距離スコアを含む。このベクトルの各要素は、libData の対応する行のスペクトル データを基準とする、テスト反射率値の距離スコアです。 |
| 1 行 1 列。1 つのスペクトル ファイル (K = 1) から読み取った基準シグネチャを含む | スカラー |
データ型: double
制限
Method を "sam"、"sid"、"jmsam"、または "ns3" として指定した場合、この関数は、パフォーマンスが既に最適化されているため、parfor ループをサポートしません。 (R2023a 以降)
アルゴリズム
"sam" 法の場合、関数 spectralMatch は、次の式を使用して SAM スコア α を計算します。
ここで、r および t は、それぞれ基準スペクトルとテスト スペクトルです。ri および ti は、それぞれベクトル r および t の i 番目の要素です。C は、ベクトル r および t の長さです。
"sid" 法の場合、関数 spectralMatch は、基準スペクトル refSpectra とテスト スペクトル testSpectra を正規化し、次の式を使用して SID 値を計算します。
q および p は、それぞれ正規化された基準スペクトルとテスト スペクトルのベクトルです。qi および pi は、それぞれベクトル q および p の i 番目の要素です。C は、ベクトル q および p の長さです。
"sidsam" 法の場合、関数 spectralMatch は、次の式を使用して SID-SAM 値を計算します。
SID は SID 値、α は SAM スコアです。
"jmsam" 法の場合、関数 spectralMatch は、次の式を使用して JMSAM スコアを計算します。
JMdistance は JM 距離、α は SAM スコアです。この手法では、次の式を使用して JM 距離を計算します。
B はバタチャリヤ距離で、以下のようになります。
μr および μt は、それぞれ基準スペクトルとテスト スペクトルの平均値です。σr および σt は、それぞれ基準スペクトルとテスト スペクトルの共分散値です。
"ns3" 法の場合、関数 spectralMatch は、次の式を使用して NS3 スコアを計算します。
AEuclidean はユークリッド距離、α は SAM スコアです。この手法では、次の式を使用してユークリッド距離を計算します。
r および t は、それぞれ基準スペクトルとテスト スペクトルのベクトルです。ri および ti は、それぞれベクトル r および t の i 番目の要素です。C は、ベクトル r および t の長さです。
バージョン履歴
R2020a で導入
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Web サイトの選択
Web サイトを選択すると、翻訳されたコンテンツにアクセスし、地域のイベントやサービスを確認できます。現在の位置情報に基づき、次のサイトの選択を推奨します:
また、以下のリストから Web サイトを選択することもできます。
最適なサイトパフォーマンスの取得方法
中国のサイト (中国語または英語) を選択することで、最適なサイトパフォーマンスが得られます。その他の国の MathWorks のサイトは、お客様の地域からのアクセスが最適化されていません。
南北アメリカ
- América Latina (Español)
- Canada (English)
- United States (English)
ヨーロッパ
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)