ns3
正規化スペクトル類似度スコアの評価
説明
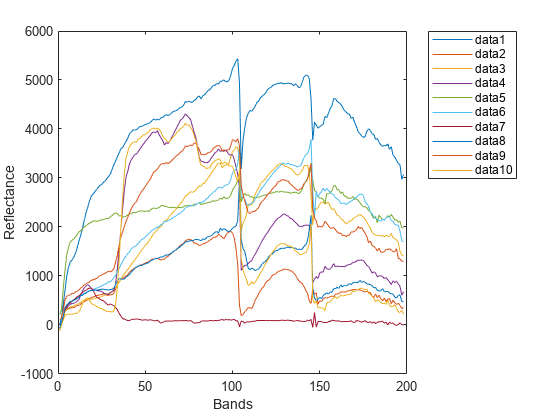
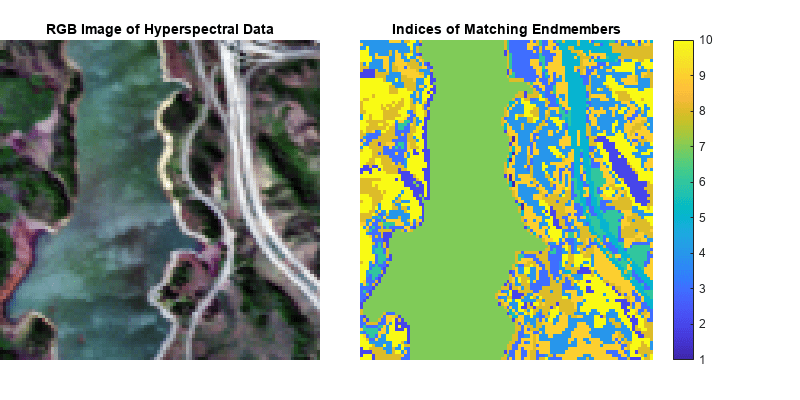
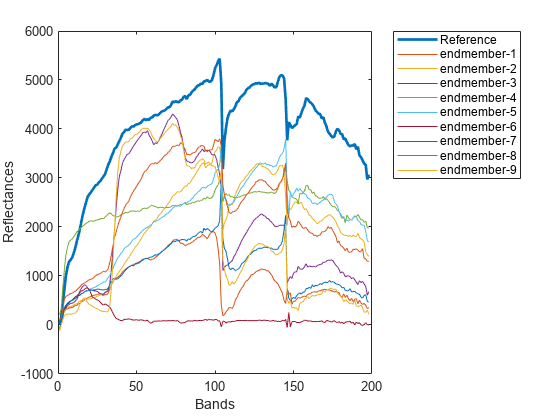
score = ns3(inputData,refSpectrum)inputData 内の各ピクセルのスペクトルと、指定した基準スペクトル refSpectrum の間のスペクトルの類似度を評価します。この構文を使用して、ハイパースペクトル データ キューブ内で異なる領域または物質を特定します。NS3 法の詳細については、詳細を参照してください。
score = ns3(testSpectrum,refSpectrum)testSpectrum と基準スペクトル refSpectrum の間のスペクトルの類似度を評価します。この構文を使用して、未知の物質のスペクトル シグネチャを基準スペクトルと比較したり、2 つのスペクトル シグネチャ間のスペクトルのばらつきを計算したりします。
メモ
この関数には、Hyperspectral Imaging Library for Image Processing Toolbox™ が必要です。Hyperspectral Imaging Library for Image Processing Toolbox はアドオン エクスプローラーからインストールできます。アドオンのインストールの詳細については、アドオンの入手と管理を参照してください。
Hyperspectral Imaging Library for Image Processing Toolbox は、MATLAB® Online™ および MATLAB Mobile™ によってサポートされないため、デスクトップの MATLAB が必要です。
例
入力引数
出力引数
制限
この関数は、パフォーマンスが既に最適化されているため、parfor ループをサポートしません。 (R2023a 以降)
詳細
参照
[1] Nidamanuri, Rama Rao, and Bernd Zbell. “Normalized Spectral Similarity Score (NS3) as an Efficient Spectral Library Searching Method for Hyperspectral Image Classification.” IEEE Journal of Selected Topics in Applied Earth Observations and Remote Sensing 4, no. 1 (March 2011): 226–40. https://doi.org/10.1109/JSTARS.2010.2086435.
バージョン履歴
R2020b で導入
参考
spectralMatch | readEcostressSig | sid | hypercube | jmsam | sidsam | sam