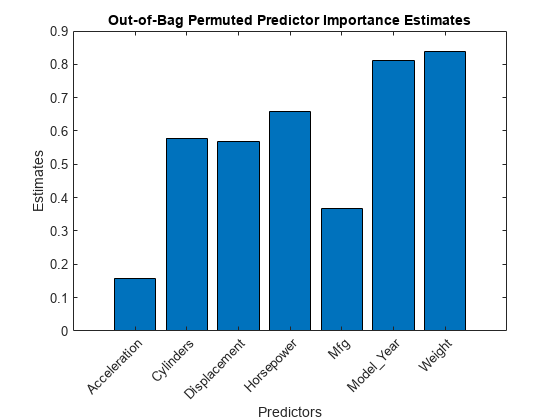
oobPermutedPredictorImportance
回帰木のランダム フォレストについての並べ替えによる out-of-bag 予測子の重要度の推定
説明
Imp = oobPermutedPredictorImportance(Mdl)Mdl を使用して、並べ替えによる out-of-bag 予測子の重要度の推定を返します。Mdl は RegressionBaggedEnsemble モデル オブジェクトでなければなりません。Imp は 1 行 p 列の数値ベクトルで、p は学習データ内の予測子変数の個数 (size(Mdl.X,2)) です。Imp( は予測子 j)Mdl.PredictorNames( の重要度です。j)
Imp = oobPermutedPredictorImportance(Mdl,Name=Value)
例
入力引数
詳細
ヒント
fitrensemble の使用によりランダム フォレストを成長させる場合、
標準 CART アルゴリズムには、相違する値が少ない分割予測子 (カテゴリカル変数など) よりも、相違する値が多い分割予測子 (連続変数など) を選択する傾向があります[3]。予測子データ セットが異種混合である場合や、一部の予測子が相違する値を他の変数よりも相対的に少なく含んでいる場合は、曲率検定または交互作用検定の指定を検討してください。
標準 CART を使用して成長させた木は、予測子変数の交互作用の影響を受けません。また、多くの無関係な予測子が存在する状況では、このような木によって重要な変数が特定される可能性は、交互作用検定を適用した場合より低くなります。このため、予測子の交互作用を考慮し、重要度変数の特定を多くの無関係な変数が存在する状況で行うには、交互作用検定を指定します[2]。
多数の予測子が学習データに含まれている場合に予測子の重要度を分析するには、アンサンブルの木学習器について関数
templateTreeのNumVariablesToSampleとして"all"を指定します。このようにしないと、重要度が過小評価されて一部の予測子が選択されない可能性があります。
詳細については、templateTree および分割予測子選択手法の選択を参照してください。
参照
[1] Breiman, L., J. Friedman, R. Olshen, and C. Stone. Classification and Regression Trees. Boca Raton, FL: CRC Press, 1984.
[2] Loh, W.Y. “Regression Trees with Unbiased Variable Selection and Interaction Detection.” Statistica Sinica, Vol. 12, 2002, pp. 361–386.
[3] Loh, W.Y. and Y.S. Shih. “Split Selection Methods for Classification Trees.” Statistica Sinica, Vol. 7, 1997, pp. 815–840.
拡張機能
バージョン履歴
R2016b で導入