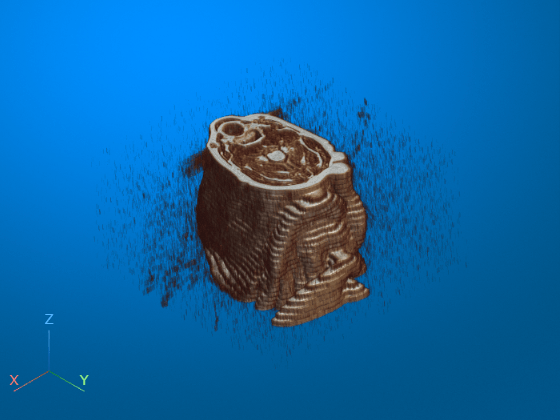
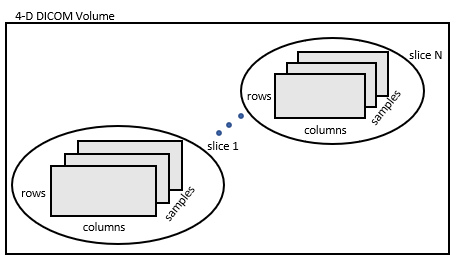
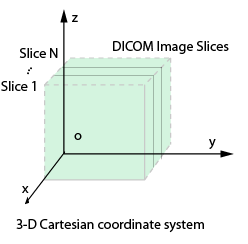
dicomreadVolume
Create 4-D volume from set of DICOM images
Syntax
Description
V = dicomreadVolume(sourceTable)sourceTable. The table must contain only one row that
specifies the metadata for a DICOM volume.
V = dicomreadVolume(sourceTable,rowname)rowname of the multirow table. Use this syntax when
sourceTable contains multiple rows.
Examples
Input Arguments
Output Arguments
Extended Capabilities
Version History
Introduced in R2017bSee Also
DICOM Browser | dicominfo | dicomread | dicomCollection | tiffreadVolume