factoran
Factor analysis
Syntax
Description
factoran computes the maximum likelihood estimate (MLE)
of the factor loadings matrix Λ in the factor analysis model
where x is a vector of observed variables, μ
is a constant vector of means, Λ is a constant
d-by-m matrix of factor loadings,
f is a vector of independent, standardized common factors,
and e is a vector of independent specific factors.
x, μ, and e each has
length d. f has length
m.
Alternatively, the factor analysis model can be specified as
where is a d-by-d diagonal matrix
of specific variances.
For the uses of factoran and its relation to pca, see Perform Factor Analysis on Exam Grades.
___ = factoran(
modifies the model fit and outputs using one or more name-value pair arguments,
for any output arguments in the previous syntaxes. For example, you can specify
that the X,m,Name,Value)X data is a covariance matrix.
Examples
Create some pseudorandom raw data.
rng default % For reproducibility n = 100; X1 = 5 + 3*rand(n,1); % Factor 1 X2 = 20 - 5*rand(n,1); % Factor 2
Create six data vectors from the raw data, and add random noise.
Y1 = 2*X1 + 3*X2 + randn(n,1); Y2 = 4*X1 + X2 + 2*randn(n,1); Y3 = X1 - X2 + 3*randn(n,1); Y4 = -2*X1 + 4*X2 + 4*randn(n,1); Y5 = 3*(X1 + X2) + 5*randn(n,1); Y6 = X1 - X2/2 + 6*randn(n,1);
Create a data matrix from the data vectors.
X = [Y1,Y2,Y3,Y4,Y5,Y6];
Extract the two factors from the noisy data matrix X using factoran. Display the outputs.
m = 2; [lambda,psi,T,stats,F] = factoran(X,m); disp(lambda)
0.8666 0.4828
0.8688 -0.0998
-0.0131 -0.5412
0.2150 0.8458
0.7040 0.2678
-0.0806 -0.2883
disp(psi)
0.0159
0.2352
0.7070
0.2385
0.4327
0.9104
disp(T)
0.8728 0.4880
0.4880 -0.8728
disp(stats)
loglike: -0.0531
dfe: 4
chisq: 5.0335
p: 0.2839
disp(F(1:10,:))
1.8845 -0.6568
-0.1714 -0.8113
-1.0534 2.0743
1.0390 -1.1784
0.4309 0.9907
-1.1823 0.6570
-0.2129 1.1898
-0.0844 -0.7421
0.5854 -1.1379
0.8279 -1.9624
View the correlation matrix of the data.
corrX = corr(X)
corrX = 6×6
1.0000 0.7047 -0.2710 0.5947 0.7391 -0.2126
0.7047 1.0000 0.0203 0.1032 0.5876 0.0289
-0.2710 0.0203 1.0000 -0.4793 -0.1495 0.1450
0.5947 0.1032 -0.4793 1.0000 0.3752 -0.2134
0.7391 0.5876 -0.1495 0.3752 1.0000 -0.2030
-0.2126 0.0289 0.1450 -0.2134 -0.2030 1.0000
Compare corrX to its corresponding values returned by factoran, lambda*lambda' + diag(psi).
C0 = lambda*lambda' + diag(psi)
C0 = 6×6
1.0000 0.7047 -0.2726 0.5946 0.7394 -0.2091
0.7047 1.0000 0.0426 0.1023 0.5849 -0.0413
-0.2726 0.0426 1.0000 -0.4605 -0.1542 0.1571
0.5946 0.1023 -0.4605 1.0000 0.3779 -0.2611
0.7394 0.5849 -0.1542 0.3779 1.0000 -0.1340
-0.2091 -0.0413 0.1571 -0.2611 -0.1340 1.0000
factoran obtains lambda and psi that correspond closely to the correlation matrix of the original data.
View the results without using rotation.
[lambda,psi,T,stats,F] = factoran(X,m,'Rotate','none'); disp(lambda)
0.9920 0.0015
0.7096 0.5111
-0.2755 0.4659
0.6004 -0.6333
0.7452 0.1098
-0.2111 0.2123
disp(psi)
0.0159
0.2352
0.7070
0.2385
0.4327
0.9104
disp(T)
1 0
0 1
disp(stats)
loglike: -0.0531
dfe: 4
chisq: 5.0335
p: 0.2839
disp(F(1:10,:))
1.3243 1.4929
-0.5456 0.6245
0.0928 -2.3246
0.3318 1.5356
0.8596 -0.6544
-0.7114 -1.1504
0.3947 -1.1424
-0.4358 0.6065
-0.0444 1.2789
-0.2350 2.1169
Compute the factors using only the covariance matrix of X.
X2 = cov(X); [lambda2,psi2,T2,stats2] = factoran(X2,m,'Xtype','covariance','Nobs',n)
lambda2 = 6×2
0.8666 0.4828
0.8688 -0.0998
-0.0131 -0.5412
0.2150 0.8458
0.7040 0.2678
-0.0806 -0.2883
psi2 = 6×1
0.0159
0.2352
0.7070
0.2385
0.4327
0.9104
T2 = 2×2
0.8728 0.4880
0.4880 -0.8728
stats2 = struct with fields:
loglike: -0.0531
dfe: 4
chisq: 5.0335
p: 0.2839
The results are the same as with the raw data, except factoran cannot compute the factor scores matrix F for covariance data.
Load the sample data.
load carbigDefine the variable matrix and remove observations with missing values.
X = [Acceleration Displacement Horsepower MPG Weight]; X = X(all(~isnan(X),2),:);
Estimate the factor loadings using a minimum mean squared error prediction for a factor analysis with two common factors. Specify to predict factor scores using a ridge regression model.
[Lambda,Psi,T,~,F] = factoran(X,2,Scores="regression")Lambda = 5×2
-0.2432 -0.8500
0.8773 0.3871
0.7618 0.5930
-0.7978 -0.2786
0.9692 0.2129
Psi = 5×1
0.2184
0.0804
0.0680
0.2859
0.0152
T = 2×2
0.9476 0.3195
0.3195 -0.9476
F = 392×2
0.4568 0.9158
0.5897 1.6438
0.2573 1.6383
0.3108 1.4154
0.2700 1.4815
1.2534 2.0922
1.1607 2.7986
1.0866 2.8106
1.2923 2.6269
0.5558 2.6860
0.3443 2.2636
0.2945 2.3158
0.6351 1.7573
-0.3769 4.1067
-0.8074 0.4072
⋮
The factor analysis model assumes that , and the elements of are the specific variances.
Calculate the estimated covariance matrix of the factor scores (F).
inv(T'*T) % Estimated covariance matrix of Fans = 2×2
1.0000 0.0000
0.0000 1.0000
Because the default rotation method for factoran is varimax, T is an orthogonal matrix, and the estimated covariance matrix of F is an identity matrix.
Calculate the estimated covariance matrix of X using the factor analysis model.
Lambda*Lambda' + diag(Psi) % Estimated covariance matrix of Xans = 5×5
1.0000 -0.5424 -0.6893 0.4309 -0.4167
-0.5424 1.0000 0.8979 -0.8078 0.9328
-0.6893 0.8979 1.0000 -0.7730 0.8647
0.4309 -0.8078 -0.7730 1.0000 -0.8326
-0.4167 0.9328 0.8647 -0.8326 1.0000
Calculate the unrotated factor loadings by multiplying by the primary axis rotation matrix.
Lambda*inv(T) % Unrotate the loadingsans = 5×2
-0.5020 0.7277
0.9550 -0.0865
0.9113 -0.3185
-0.8450 0.0091
0.9865 0.1079
Calculate the unrotated factor scores by multiplying F by T'.
F*T' % Unrotate the factor scoresans = 392×2
0.7255 -0.7219
1.0840 -1.3692
0.7673 -1.4702
0.7467 -1.2419
0.7292 -1.3176
1.8562 -1.5820
1.9940 -2.2811
1.9276 -2.3161
2.0638 -2.0764
1.3848 -2.3676
1.0494 -2.0349
1.0190 -2.1003
1.1633 -1.4622
0.9549 -4.0119
-0.6349 -0.6438
⋮
Create a biplot of two factors.
biplot(Lambda,LineWidth=2,MarkerSize=20)
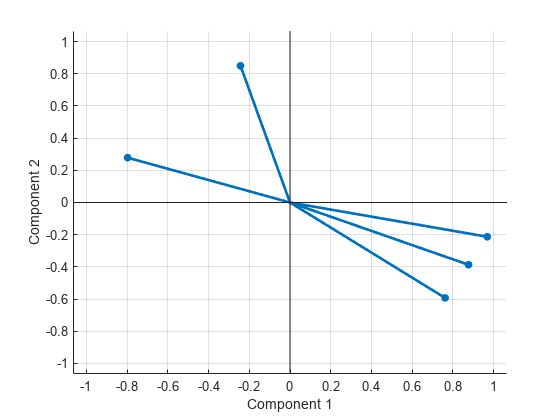
Estimate the factor loadings using the covariance (or correlation) matrix.
[Lambda,Psi,T] = factoran(cov(X),2,Xtype="covariance")Lambda = 5×2
-0.2432 -0.8500
0.8773 0.3871
0.7618 0.5930
-0.7978 -0.2786
0.9692 0.2129
Psi = 5×1
0.2184
0.0804
0.0680
0.2859
0.0152
T = 2×2
0.9476 0.3195
0.3195 -0.9476
You can also use corrcoef(X) instead of cov(X) to create the data for factoran.
Although the estimates are the same, the use of a covariance matrix rather than raw data prevents you from requesting the scores or significance level.
Rotate the factors and scores using the promax method.
[Lambda,Psi,T,stats,F] = factoran(X,2,Rotate="promax"); inv(T'*T) % Estimated covariance matrix of F, no longer eye(2)
ans = 2×2
1.0000 -0.6391
-0.6391 1.0000
Lambda*inv(T'*T)*Lambda'+diag(Psi) % Estimated covariance matrix of Xans = 5×5
1.0000 -0.5424 -0.6893 0.4309 -0.4167
-0.5424 1.0000 0.8979 -0.8078 0.9328
-0.6893 0.8979 1.0000 -0.7730 0.8647
0.4309 -0.8078 -0.7730 1.0000 -0.8326
-0.4167 0.9328 0.8647 -0.8326 1.0000
Plot the unrotated variables with oblique axes superimposed.
invT = inv(T); Lambda0 = Lambda*invT; figure() line([-invT(1,1) invT(1,1) NaN -invT(2,1) invT(2,1)], ... [-invT(1,2) invT(1,2) NaN -invT(2,2) invT(2,2)], ... Color="b",LineWidth=2) grid on hold on biplot(Lambda0,LineWidth=2,MarkerSize=20) xlabel("Loadings for Unrotated Factor 1") ylabel("Loadings for Unrotated Factor 2")
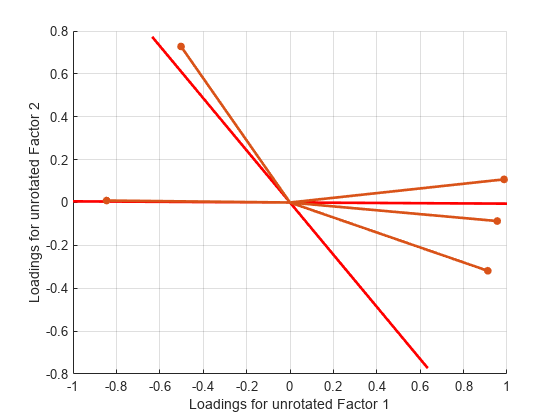
Plot the rotated variables against the oblique axes.
figure() biplot(Lambda,LineWidth=2,MarkerSize=20)
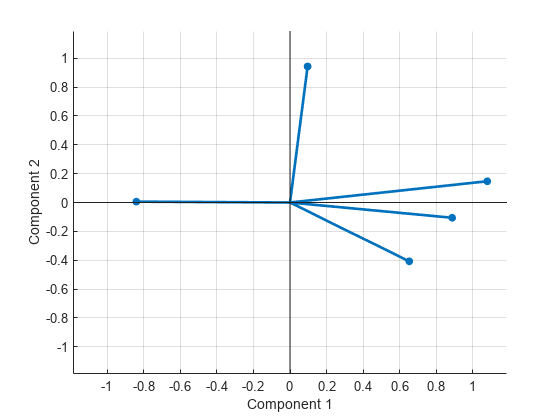
Input Arguments
Data, specified as an n-by-d
matrix, where each row is an observation of d
variables.
Data Types: double
Number of common factors, specified as a positive integer. The
maximum allowable value of m must satisfy the
inequality (d –
m)2 –
d – m ≥
0, where d is the number of
variables in X. This condition ensures that the
error degrees of freedom value is nonnegative (see stats).
Example: 3
Data Types: double
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Example: lambda = factoran(X,m,'Start',10,'Scores','Thomson')
specifies to use a starting point for specific variances of 10 and the Thomson
method for predicting factor scores.
Input data type of X, specified as the
comma-separated pair consisting of 'Xtype' and
one of the following:
'data'—Xis raw data.'covariance'—Xis a positive definite covariance or correlation matrix.
Example: 'Xtype','covariance'
Data Types: char | string
Method for predicting factor scores, specified as the
comma-separated pair consisting of 'Scores'
and one of the following:
'wls'or the equivalent'Bartlett'— Weighted least-squares estimate treatingFas fixed'regression'or the equivalent'Thomson'— Minimum mean squared error prediction that is equivalent to a ridge regression
Example: 'Scores','regression'
Data Types: char | string
Starting point for the specific variances psi
in the maximum likelihood optimization, specified as the
comma-separated pair consisting of 'Start' and
one of the following:
'Rsquared'— Chooses the starting vector as a scale factor timesdiag(inv(corrcoef(X)))(default). For examples, see Jöreskog [2].'random'— Choosesduniformly distributed values on the interval [0,1].Positive integer — Performs the given number of maximum likelihood fits, each initialized in the same way as
'random'.factoranreturns the fit with the highest likelihood.Matrix with
drows — Performs one maximum likelihood fit for each column of the specified matrix.factoraninitializes theith optimization with the values from theith column.
Example: 'Start',5
Data Types: double | char | string
Method used to rotate factor loadings and scores, specified as
the comma-separated pair consisting of
'Rotate' and one of the values in the
following table. You can control the rotation by specifying
additional name-value pair arguments of the
rotatefactors function, as described in
the table. For details, see rotatefactors.
| Value | Description |
|---|---|
| Performs no rotation |
| Special case of the
|
| Orthogonal rotation that maximizes a
criterion based on the variance of the loadings.
Use the |
| Special case of the orthomax
rotation. Use the |
| Performs either an oblique rotation
(the default) or an orthogonal rotation to best
match a specified pattern matrix. Use the
|
| Performs either an oblique rotation
(the default) or an orthogonal rotation to best
match a specified target matrix in the
least-squares sense. Use the
|
| Performs an oblique procrustes
rotation to a target matrix determined by
|
| Special case of the
|
| Special case of the
|
function handle | Function handle to a rotation function of the form [B,T] = myrotation(A,...) where
Use the
|
Example: [lambda,psi,T] =
factoran(X,m,'Rotate','promax','power',5,'maxit',100)
Data Types: char | string | function_handle
Lower bound for the psi argument during
maximum likelihood optimization, specified as the comma-separated
pair consisting of 'Delta' and a scalar value
between 0 and 1 (0 < Delta < 1).
Example: 0.02
Data Types: double
Options for the maximum likelihood optimization, specified as
the comma-separated pair consisting of
'OptimOpts' and a structure created by
statset. You can enter
statset('factoran') for the list of
options, which are also described in the following table.
Field Name (statset
argument) | Meaning | Value {default} |
|---|---|---|
'Display' | Amount of information displayed by the algorithm |
|
MaxFunEvals | Maximum number of objective function evaluations allowed | Positive integer,
{400} |
MaxIter | Maximum number of iterations allowed | Positive integer,
{100} |
TolFun | Termination tolerance for the
objective function value. The solver stops when
successive function values are less than
| Positive scalar,
{1e-8} |
TolX | Termination tolerance for the
parameters. The solver stops when successive
parameter values are less than
| Positive scalar,
{1e-8} |
Example: statset('Display','iter')
Data Types: struct
Output Arguments
Factor loadings, returned as a
d-by-m matrix.
d is the number of columns of the data matrix
X, and m is the second
input argument of factoran.
The (i,j)th element of lambda
is the coefficient, or loading, of the jth factor
for the ith variable. By default,
factoran calls the function
rotatefactors to rotate the estimated factor
loadings using the 'varimax' option. For
information about rotation, see Rotation of Factor Loadings and Scores.
Specific variances, returned as a
d-by-1 vector.
d is the number of columns of the data matrix
X. The entries of psi are
maximum likelihood estimates.
Factor loadings rotation, returned as an
m-by-m matrix.
m is the second input argument of
factoran. For information about rotation,
see Rotation of Factor Loadings and Scores.
Information about the common factors, returned as a structure.
stats contains information relating to the
null hypothesis H0 that the number of common
factors is m
[3].
stats contains the following fields.
| Field | Description |
|---|---|
loglike | Maximized loglikelihood value |
dfe | Error degrees of freedom = (( |
chisq | Approximate chi-square statistic for the null hypothesis |
p | Right-tail significance level for the null hypothesis |
factoran does not compute the
chisq and p fields unless
dfe is positive and all the specific variance
estimates in psi are positive (see Heywood Case).
If X is a covariance matrix and you want
factoran to compute the
chisq and p fields, then
you must also specify the 'Nobs' name-value pair
argument.
Factor scores, also called predictions of the common factors,
returned as an n-by-m matrix.
n is the number of rows in the data matrix
X, and m is the second
input argument of factoran.
Note
If X is a covariance matrix
(Xtype =
'covariance'),
factoran cannot compute
F.
factoran rotates F using the
same criterion as for lambda. For information about
rotation, see Rotation of Factor Loadings and Scores.
More About
If elements of psi are equal to the value
of the Delta parameter (that is, they are essentially zero),
the fit is known as a Heywood case, and interpretation of the resulting
estimates is problematic. In particular, there can be multiple local maxima of
the likelihood, each with different estimates of the loadings and the specific
variances. Heywood cases can indicate overfitting (m is too
large), but can also be the result of underfitting.
Unless you explicitly specify no rotation using the
'Rotate' name-value pair argument,
factoran rotates the estimated factor loadings
lambda and the factor scores F.
The output matrix T is used to rotate the loadings, that
is, lambda = lambda0*T, where
lambda0 is the initial (unrotated) MLE of the loadings.
T is an orthogonal matrix for orthogonal rotations,
and the identity matrix for no rotation. The inverse of T is
known as the primary axis rotation matrix, whereas T itself
is related to the reference axis rotation matrix. For orthogonal rotations, the
two are identical.
factoran computes factor scores that have been rotated by
inv(T'), that is,
F = F0 * inv(T'), where
F0 contains the unrotated predictions. The estimated
covariance of F is inv(T'*T), which is the
identity matrix for orthogonal or no rotation. Rotation of factor loadings and
scores is an attempt to create a structure that is easier to interpret in the
loadings matrix after maximum likelihood estimation.
The syntax for passing additional arguments to a user-defined rotation function is:
[Lambda,Psi,T] = ... factoran(X,2,'Rotate',@myrotation,'UserArgs',1,'two');
References
[1] Harman, Harry Horace. Modern Factor Analysis. 3rd Ed. Chicago: University of Chicago Press, 1976.
[2] Jöreskog, K. G. “Some Contributions to Maximum Likelihood Factor Analysis.” Psychometrika 32, no. 4 (December 1967): 443–82. https://doi.org/10.1007/BF02289658
[3] Krzanowski, W. J. Principles of Multivariate Analysis: A User's Perspective. New York: Oxford University Press, 1988.
[4] Lawley, D. N., and A. E. Maxwell. Factor Analysis as a Statistical Method. 2nd Ed. New York: American Elsevier Publishing Co., 1971.
Extended Capabilities
pcacov and factoran
do not work directly on tall arrays. Instead, use C = gather(cov(X)) to
compute the covariance matrix of a tall array. Then, you can use pcacov or
factoran to work on the in-memory covariance matrix. Alternatively, you
can use pca directly on a tall array.

For more information, see Tall Arrays for Out-of-Memory Data.
Version History
Introduced before R2006a
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Select a Web Site
Choose a web site to get translated content where available and see local events and offers. Based on your location, we recommend that you select: .
You can also select a web site from the following list
How to Get Best Site Performance
Select the China site (in Chinese or English) for best site performance. Other MathWorks country sites are not optimized for visits from your location.
Americas
- América Latina (Español)
- Canada (English)
- United States (English)
Europe
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)