margin
ガウス カーネル分類モデルの分類マージン
説明
例
ionosphere データ セットを読み込みます。このデータ セットには、レーダー反射についての 34 個の予測子と、不良 ('b') または良好 ('g') という 351 個の二項反応が含まれています。
load ionosphereデータ セットを学習セットとテスト セットに分割します。テスト セット用に 30% のホールドアウト標本を指定します。
rng('default') % For reproducibility Partition = cvpartition(Y,'Holdout',0.30); trainingInds = training(Partition); % Indices for the training set testInds = test(Partition); % Indices for the test set
学習セットを使用してバイナリ カーネル分類モデルに学習をさせます。
Mdl = fitckernel(X(trainingInds,:),Y(trainingInds));
学習セットのマージンとテストセットのマージンを推定します。
mTrain = margin(Mdl,X(trainingInds,:),Y(trainingInds)); mTest = margin(Mdl,X(testInds,:),Y(testInds));
箱ひげ図を使用して、両方のマージンのセットをプロットします。
boxplot([mTrain; mTest],[zeros(size(mTrain,1),1); ones(size(mTest,1),1)], ... 'Labels',{'Training set','Test set'}); title('Training-Set and Test-Set Margins')
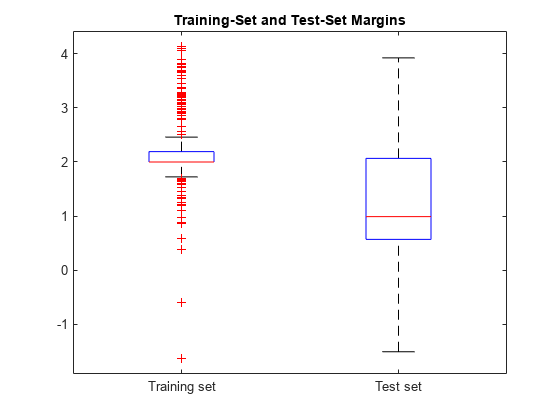
学習セットのマージン分布はテスト セットのマージン分布より高い位置にあります。
複数のモデルによるテストセットのマージンを比較することにより、特徴選択を実行します。この基準のみに基づくと、マージンが大きい方が分類器として優れています。
ionosphere データ セットを読み込みます。このデータ セットには、レーダー反射についての 34 個の予測子と、不良 ('b') または良好 ('g') という 351 個の二項反応が含まれています。
load ionosphereデータ セットを学習セットとテスト セットに分割します。テスト セット用に 15% のホールドアウト標本を指定します。
rng('default') % For reproducibility Partition = cvpartition(Y,'Holdout',0.15); trainingInds = training(Partition); % Indices for the training set XTrain = X(trainingInds,:); YTrain = Y(trainingInds); testInds = test(Partition); % Indices for the test set XTest = X(testInds,:); YTest = Y(testInds);
予測子変数の 10% を無作為に選択します。
p = size(X,2); % Number of predictors
idxPart = randsample(p,ceil(0.1*p));2 つのバイナリ カーネル分類モデルに学習をさせます。1 つではすべての予測子を、もう 1 つではランダムな 10% の予測子を使用します。
Mdl = fitckernel(XTrain,YTrain); PMdl = fitckernel(XTrain(:,idxPart),YTrain);
Mdl および PMdl は ClassificationKernel モデルです。
各分類器についてテストセットのマージンを推定します。
fullMargins = margin(Mdl,XTest,YTest); partMargins = margin(PMdl,XTest(:,idxPart),YTest);
箱ひげ図を使用して、マージン セットの分布をプロットします。
boxplot([fullMargins partMargins], ... 'Labels',{'All Predictors','10% of the Predictors'}); title('Test-Set Margins')
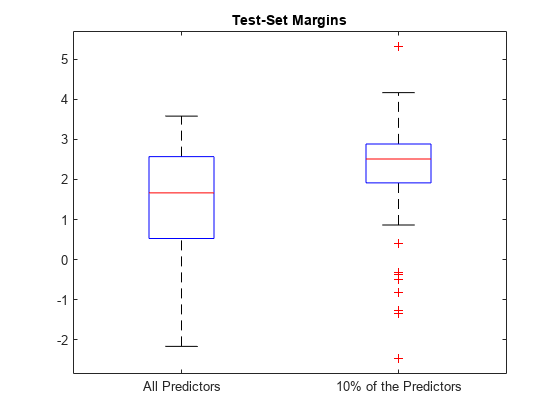
PMdl のマージン分布は Mdl のマージン分布より高い位置にあります。したがって、PMdl モデルの方が優れた分類器です。
入力引数
バイナリ カーネル分類モデル。ClassificationKernel モデル オブジェクトを指定します。ClassificationKernel モデル オブジェクトは、fitckernel を使用して作成できます。
クラス ラベル。categorical 配列、文字配列、string 配列、logical ベクトル、数値ベクトル、または文字ベクトルの cell 配列を指定します。
データ型: categorical | char | string | logical | single | double | cell
モデルを学習させるために使用する標本データ。table として指定します。Tbl の各行は 1 つの観測値に、各列は 1 つの予測子変数に対応します。必要に応じて、応答変数用および観測値の重み用の追加列を Tbl に含めることができます。Tbl には、Mdl を学習させるために使用したすべての予測子が含まれていなければなりません。文字ベクトルの cell 配列ではない cell 配列と複数列の変数は使用できません。
Mdl を学習させるために使用した応答変数が Tbl に含まれている場合、ResponseVarName または Y を指定する必要はありません。
テーブルに格納されている標本データを使用して Mdl の学習を行った場合、margin の入力データもテーブルに格納されていなければなりません。
応答変数の名前。Tbl 内の変数の名前で指定します。Mdl を学習させるために使用した応答変数が Tbl に含まれている場合、ResponseVarName を指定する必要はありません。
ResponseVarName を指定する場合は、文字ベクトルまたは string スカラーとして指定しなければなりません。たとえば、応答変数が Tbl.Y として格納されている場合、ResponseVarName として 'Y' を指定します。それ以外の場合、Tbl の列は Tbl.Y を含めてすべて予測子として扱われます。
応答変数は、categorical 配列、文字配列、string 配列、logical ベクトル、数値ベクトル、または文字ベクトルの cell 配列でなければなりません。応答変数が文字配列の場合、各要素は配列の 1 つの行に対応しなければなりません。
データ型: char | string
詳細
バイナリ分類の "分類マージン" は、各観測値における真のクラスの分類スコアと偽のクラスの分類スコアの差です。
このソフトウェアでは、バイナリ分類の分類マージンは次のように定義されます。
x は観測値です。x の真のラベルが陽性クラスである場合、y は 1、それ以外の場合は –1 です。f(x) は観測値 x についての陽性クラスの分類スコアです。一般的には、分類マージンは m = yf(x) と定義されています。
各マージンのスケールが同じである場合、マージンを分類の信頼尺度として使用できます。複数の分類器の中で、マージンが大きい分類器の方が優れています。
カーネル分類モデルの場合、観測値 x (行列ベクトル) を陽性クラスに分類する生の "分類スコア" は次のように定義されます。
は特徴量を拡張するための観測値の変換です。
β は推定された係数の列ベクトルです。
b は推定されたスカラー バイアスです。
x を陰性クラスに分類する生の分類スコアは −f(x) です。このソフトウェアでは、スコアが正になるクラスに観測値が分類されます。
カーネル分類モデルがロジスティック回帰学習器から構成されている場合、'logit' スコア変換が生の分類スコアに適用されます (ScoreTransform を参照)。
拡張機能
margin 関数は、tall 配列を次の使用上の注意事項および制限事項付きでサポートします。
marginは talltableデータをサポートしていません。
詳細は、tall 配列を参照してください。
この関数は、GPU 配列を完全にサポートします。詳細は、GPU での MATLAB 関数の実行 (Parallel Computing Toolbox)を参照してください。
バージョン履歴
R2017b で導入margin は GPU 配列を完全にサポートします。
参考
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
Web サイトの選択
Web サイトを選択すると、翻訳されたコンテンツにアクセスし、地域のイベントやサービスを確認できます。現在の位置情報に基づき、次のサイトの選択を推奨します:
また、以下のリストから Web サイトを選択することもできます。
最適なサイトパフォーマンスの取得方法
中国のサイト (中国語または英語) を選択することで、最適なサイトパフォーマンスが得られます。その他の国の MathWorks のサイトは、お客様の地域からのアクセスが最適化されていません。
南北アメリカ
- América Latina (Español)
- Canada (English)
- United States (English)
ヨーロッパ
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)