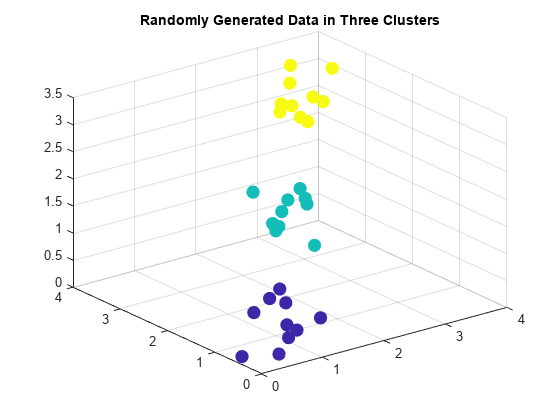
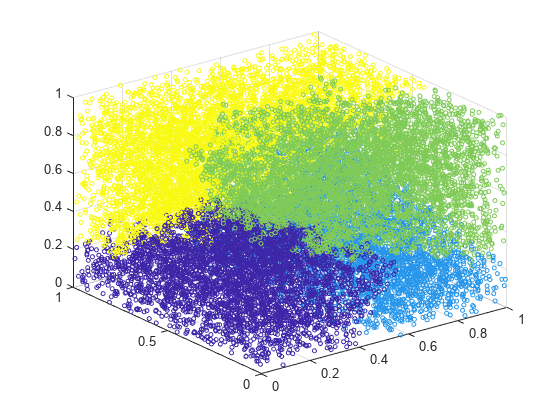
clusterdata
データから凝集型クラスターを構築
構文
説明
T = clusterdata(___,Name=Value)clusterdata(X,MaxClust=5,Depth=3) と指定します。
例
入力引数
名前と値の引数
出力引数
ヒント
Linkageが"centroid"または"median"である場合、linkageは非単調なクラスター ツリーを作成する可能性があります。このようになるのは、2 つのクラスター r および s の結合から 3 番目のクラスターまでの距離が r と s の間の距離より小さい場合です。この場合、既定の方向に描画したデンドログラムでは、葉からルート ノードへのパスは下向きになります。これを回避するには、Linkageについて別の値を指定します。次の図に非単調のクラスター ツリーを示します。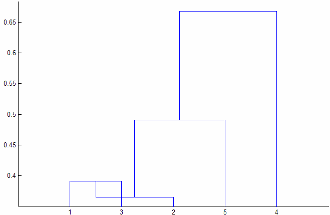
この場合、クラスター 1 とクラスター 3 が結合され新しいクラスターになっていますが、この新しいクラスターとクラスター 2 の間の距離はクラスター 1 とクラスター 3 の間の距離よりも短くなっています。
アルゴリズム
オプションの名前と値の引数をいずれも指定しない場合、clusterdata 関数は次の手順を実行します。
pdistを使用して、X内の観測値のペア間のユークリッド距離が含まれているベクトルを作成します。Y =pdist(X,"euclidean")"single"法でlinkageを使用してクラスター間の最短距離を計算することにより、凝集型階層クラスター ツリーをYから作成します。Z =linkage(Y,"single")cutoffを指定すると、clusterdata関数は、不整合値がcutoff未満であればclusterを使用してZからクラスターを定義します。T=cluster(Z,Cutoff=cutoff)maxclustを指定すると、clusterdata関数は、clusterを使用してZから最大maxclust個のクラスターを検出します。クラスターを定義する基準には"distance"を使用します。T= cluster(Z,MaxClust=maxclust)
代替機能
階層クラスター ツリー Z (入力データ行列 X に対する関数 linkage の出力) がある場合、cluster を使用すると、Z に対して凝集型クラスタリングを実行して、X 内の各観測値 (行) のクラスター割り当てを取得できます。