phytree
Data structure containing phylogenetic tree
Description
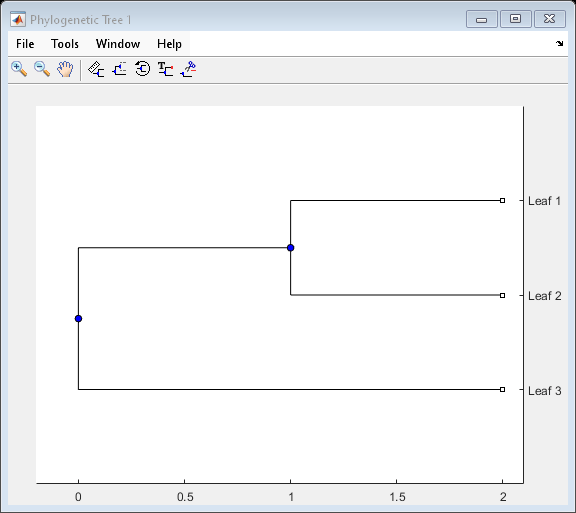
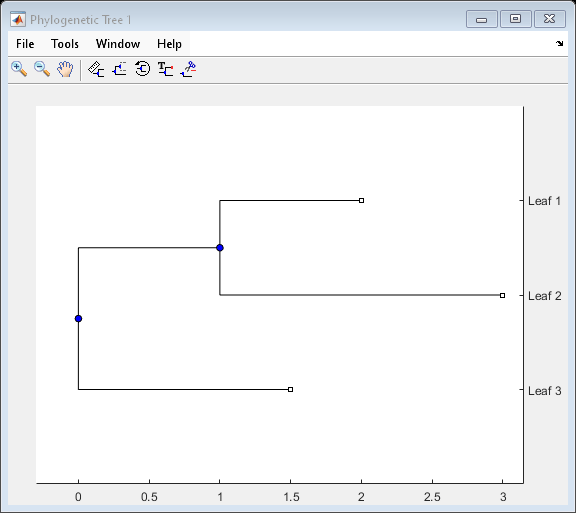
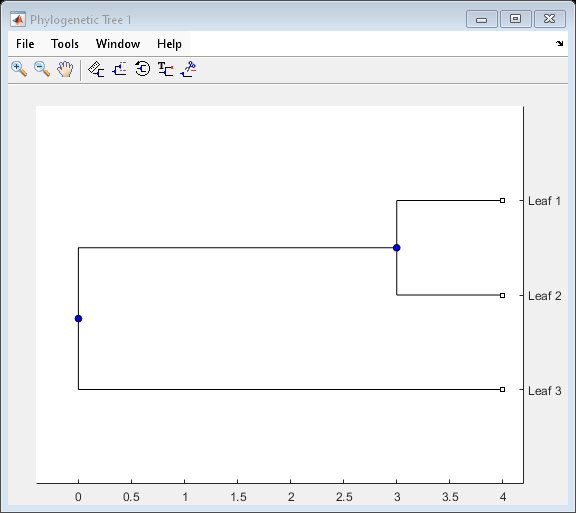
A phytree object is a data structure containing a phylogenetic
tree. Phylogenetic trees are binary rooted trees, which means that each branch is the parent
of two other branches, two leaves, or one branch and one leaf. A phytree
object can be ultrametric or nonultrametric.
Creation
Syntax
Description
tree = phytree(B)
tree = phytree(BC)BC(:,[1 2]) and branch coordinates in BC(:,3).
Same as phytree(B,C).
tree = phytree(___,N)
tree = phytree
Input Arguments
Properties
Object Functions
cluster | Validate clusters in phylogenetic tree |
get | Retrieve information about phylogenetic tree object |
getbyname | Branches and leaves from phytree object |
getcanonical | Calculate canonical form of phylogenetic tree |
getmatrix | Convert phytree object into relationship matrix |
getnewickstr | Create Newick-formatted character vector |
pdist | Calculate pairwise patristic distances in phytree object |
plot | Draw phylogenetic tree |
prune | Remove branch nodes from phylogenetic tree |
reorder | Reorder leaves of phylogenetic tree |
reroot | Change root of phylogenetic tree |
select | Select tree branches and leaves in phytree object |
subtree | Extract phylogenetic subtree |
view | View phylogenetic tree |
weights | Calculate weights for phylogenetic tree |
Examples
Version History
Introduced in R2006b
See Also
Functions
phytreeread|phytreeviewer|phytreewrite|seqlinkage|seqneighjoin|seqpdist|cluster|get|getbyname|getcanonical|getmatrix|getnewickstr|pdist|plot|prune|reroot|select|subtree|view|weights