AllenAtlasTopDown
Allen Atlas Top-Down Viewer
This package provides code to generate a top-down view of the Allen Atlas from the full Common Coordinate Framework (CCF) and visualize it interactively in stereotaxic coordinates expressed in mm with respect to bregma. There are two functions for plotting the view as well as all code needed to generate the plots from scratch using the CCF.
Installation
The easiest way to to install and stay up to date is directly via MATLAB: Go to "APPS" in the ribbon then select "Get More Apps" and search for AllenAtlasTopDown to install.
Alternatively you can download the Zip archive or clone the repo then add the code directory to your MATLAB path.
No need to "Add with Subfolders".
This package requires the Image Processing Toolbox.
Usage
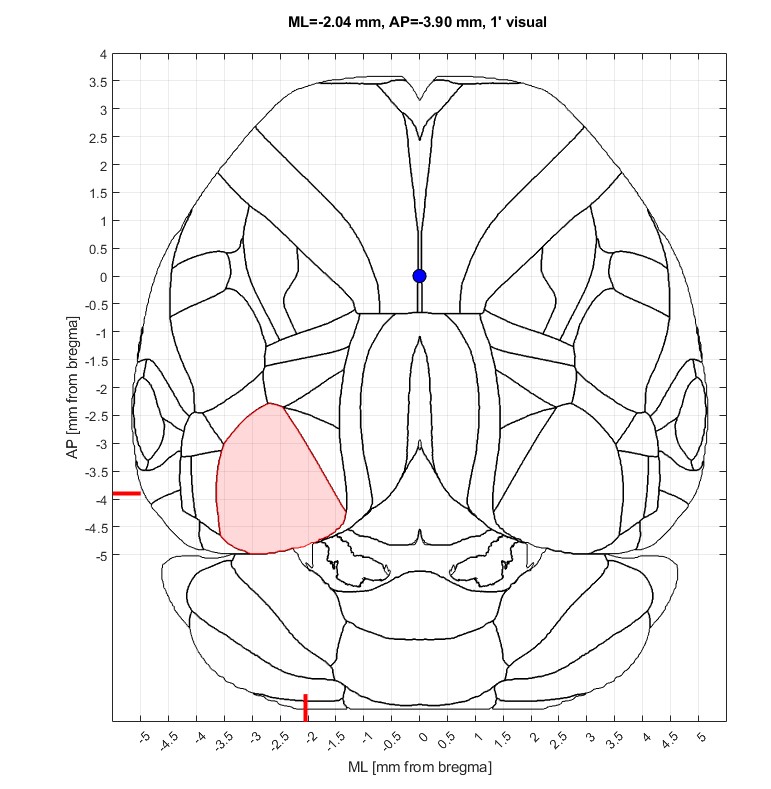
To launch an interactive top-down viewer:
aratopdown.area_highlighterTo show a static image with labels:
aratopdown.draw_top_down_ccfTo modify details of the plotted areas
The area borders are generated from the full CCF volume. Following installation of the requirements described below, the borders can be re-generated as follows.
>> t = aratopdown.atlas.build_topdown
t =
struct with fields:
bregma: [540 44 570]
dorsal_brain_areas: [43×1 struct]
whole_brain: [1×1 struct]
plot_areas: [50×1 uint16]
top_down_annotation: [1×1 struct]
% Then plotted as above
>> aratopdown.draw_top_down_ccf(t);
>> aratopdown.area_highlighter(t)Requirements for building the top-down view from the Allen CCF volume
These requirements are for re-building the area borders and are not needed for running the two
visualisation functions.
To build the top-down view you should download the Allen CCF mouse atlas from https://osf.io/fv7ed/.
You will need the files structure_tree_safe_2017.csv and annotation_volume_10um_by_index.npy
These files are a re-formatted version of the original atlas, which has been processed with this script)
Place the files somewhere in your MATLAB path.
Download/clone npy-matlab and add to your MATLAB path.
Mouse CCF scaling, rotation, and bregma notes
Bregma has been approximated in AP by matching the Paxinos atlas slice at AP=0 to the CCF, the ML position is the midline, and the DV position is a very rough approximation from matching an MRI image (this DV coordinate shouldn't be used - all actual coordinates should be measured from the brain surface for better accuracy)
The CCF is slightly stretched in the DV axis (because it's based on a single mouse with an unusually tall brain), currently estimated here as 94.3%. This stretch has not been taken into account in this package.
The CCF AP rotation is arbitrary with reference to the skull. It is estimated that this angle is 5 degrees nose-down (from https://www.biorxiv.org/content/10.1101/2022.05.09.491042v3). Since this will have negligable impact on the top-down view, the brain is not un-tilted in this implementation.
Acknowledgements
This code is derived from work done by Andy Peters in his neuropixels_trajectory_explorer. Furthermore, Andy kindly provided an additional script that demonstrated how to elegantly get the top-down ARA view in coordinates with respect to bregma. Some text from this README is taken from the neuropixels_trajectory_explorer README.
More info
Changelog
- 2023-01-05: Initial commit. Minor bugfixes and improvements.
引用
Rob Campbell (2024). AllenAtlasTopDown (https://github.com/Zapit-Optostim/AllenAtlasTopDown/releases/tag/v0.1.4), GitHub. に取得済み.
MATLAB リリースの互換性
プラットフォームの互換性
Windows macOS Linuxカテゴリ
タグ
Community Treasure Hunt
Find the treasures in MATLAB Central and discover how the community can help you!
Start Hunting!code/+aratopdown
code/+aratopdown/+atlas
| バージョン | 公開済み | リリース ノート | |
|---|---|---|---|
| 0.1.4.0 | See release notes for this release on GitHub: https://github.com/Zapit-Optostim/AllenAtlasTopDown/releases/tag/v0.1.4 |
||
| 0.1.3 | See release notes for this release on GitHub: https://github.com/Zapit-Optostim/AllenAtlasTopDown/releases/tag/v0.1.3 |
||
| 0.1.2 | See release notes for this release on GitHub: https://github.com/Zapit-Optostim/AllenAtlasTopDown/releases/tag/v0.1.2 |