Estimate Threshold-Switching Dynamic Regression Models
This example shows how to use the estimate function to fit threshold-switching dynamic regression models to simulated data and to tune the estimation procedure. The example estimates a self-exciting threshold autoregressive model (SETAR), a TAR model with an exogenous threshold variable, and a self-exciting smooth TAR model (SESTAR) containing a regression component.
Threshold-switching dynamic regression model estimation, performed by estimate, is an iterative two-step procedure:
Conduct a constrained, nonlinear search over threshold transition levels and rates.
Solve a linear least-squares problem for submodel parameters.
The resulting estimates of parameter standard errors, data loglikelihood, and model residuals are conditional on the optimal threshold parameters computed by the search.
Simulate Data from SETAR DGP
Consider a data-generating process (DGP) for the following simple univariate, mean-switching, SETAR model:
The discrete, endogenous threshold transition is at mid-level 1.5.
is the model-wide process variance.
The state 1 submodel is , where .
The state 2 submodel is , where .
Create a SETAR model that represents the DGP by following this procedure:
Create the threshold transition by using the
thresholdfunction. The resultingthresholdobject represents all aspects of the threshold transition except for whether the associated threshold variable is endogenous or exogenous to the threshold-switching model system.Create each univariate submodel by using the
arimafunction. The resultingarimaobjects each characterize the dynamics of the system within their respective states. Concatenate the model objects in a vector.Create the threshold-switching model by passing the threshold transitions object and state submodel vector to the
tsVARfunction. Specify the model-wide covariance by using theCovariancename-value argument. The resultingtsVARobject represents all aspects of the threshold-switching model for whether the associated threshold variable is endogenous or exogenous to its system. Object functions of thetsVARobject require knowledge of the threshold variable type (the default is endogenous or self-exciting for all object functions, with a delay of 1 time step).
midlvl = 1.5; ttDGP = threshold(midlvl); mdl1DGP = arima(Constant=0,AR=0.2); mdl2DGP = arima(Constant=2,AR=0.4); MdlDGP = tsVAR(ttDGP,[mdl1DGP mdl2DGP],Covariance=1);
MdlDGP is a tsVAR object that represents the DGP. It is fully specified because all parameters are set to known values.
Generate a random path of length 500 from the SETAR DGP by using the simulate object function. Plot the threshold transition and the path.
seed = 0; rng(seed) % For reproducibility T = 500; % Sample size Y = simulate(MdlDGP,T); % SETAR path figure ttplot(ttDGP,Data=Y)

The plot shows the self-exciting response variable sampling states on either side of the threshold level. In practice, without knowledge of the DGP, a plot of empirical data in Y informs a reasonable choice of the initial threshold level, which is estimate requires.
Create Model Template for Estimation
To estimate a SETAR model of the data, create the following variables:
A model template for estimation, which is a partially specified
tsVARobject that indicates the structure of the target model withNaNvalues for estimable parametersA threshold transition to initialize the search, which is a fully specified
thresholdobject. This example uses1as the initial mid-level.
tt = threshold(NaN); mdl1 = arima(Constant=NaN,AR=NaN); mdl2 = arima(Constant=NaN,AR=NaN); Mdl = tsVAR(tt,[mdl1 mdl2],Covariance=NaN); t0 = 1; % Initial mid-level tt0 = threshold(t0,Type="discrete");
Fit SETAR Model to Data
Discrete transitions pose a unique estimation challenge because they fail to disambiguate levels that fall between threshold variable data, which produces discontinuous objective functions that are difficult to optimize. To address this problem, estimate treats discrete transition levels as high-rate logistic transitions before it converts to discrete levels and discards the rates at estimation termination [1]. The MaxRate name-value argument specifies the upper bound for the rates estimate uses for estimation.
This code shows the internal representation of the discrete transition during estimation by plotting a logistic threshold transition at mid-level 1.5 with rate 10.
maxRate = 10; ttEst = threshold(midlvl,Type="logistic",Rates=maxRate); figure ttplot(ttEst,Data=Y) colorbar("off")

The logistic transition combines the two submodels in a weighted mixture model that is responsive to continuous changes in level during the threshold search procedure. estimate automatically implements the transformation from a discrete to a logistic representation at the beginning of estimation, and then it reverse-transforms the resulting threshold transition at estimation termination.
Estimate the SETAR model by passing the model template Mdl, initial threshold transition tt0, and simulated path Y to estimate. Set the maximum transition rate to 10. By default, the threshold variable is endogenous (Type="endogenous") with a delay of one time step (Delay=1).
EstMdl = estimate(Mdl,tt0,Y,MaxRate=maxRate);
EstMdl is a fully specified tsVAR object representing the estimated SETAR model with a discrete threshold transition. estimate uses the maximum rate maxRate only internally because it represents the discrete transition as logistic to overcome numerical problems during optimization. To tune the estimation procedure for any threshold transition type, you can iterate by adjusting MaxRate and calling estimate.
Inspect Estimation Results
Display the estimated threshold transition Switch, stored in the estimated model object, by using dot notation.
EstTT = EstMdl.Switch
EstTT =
threshold with properties:
Type: 'discrete'
Levels: 1.5233
Rates: []
StateNames: ["1" "2"]
NumStates: 2
Display the estimated submodels.
EstMdl.Submodels
ans =
2×1 varm array with properties:
NumSeries
P
Constant
AR
Beta
Trend
Covariance
SeriesNames
Description
Observe that estimate returns a vector of varm model objects representing the estimated univariate models, although the model template Mdl specifies a vector of arima model objects.
Visually compare the estimated parameters to those of the DGP.
EstMdlParams = [EstMdl.Switch.Levels; ... EstMdl.Submodels(1).Constant; ... EstMdl.Submodels(2).Constant; ... EstMdl.Submodels(1).AR{:}; ... EstMdl.Submodels(2).AR{:}; ... EstMdl.Covariance]; DGPParams = [MdlDGP.Switch.Levels; ... MdlDGP.Submodels(1).Constant; ... MdlDGP.Submodels(2).Constant; ... MdlDGP.Submodels(1).AR{:}; ... MdlDGP.Submodels(2).AR{:}; ... MdlDGP.Covariance]; table(DGPParams,EstMdlParams)
ans=6×2 table
DGPParams EstMdlParams
_________ ____________
1.5 1.5233
0 0.034171
2 1.8688
0.2 0.14564
0.4 0.41827
1 1.0406
The estimation results, with sufficient sample size and a limited number of parameters, approximate values of the DGP.
Summarize the estimation results.
summarize(EstMdl)
Description
1-Dimensional tsVAR Model with 2 Submodels
Switch
Transition Type: discrete
Estimated Levels: 1.523
Fit
Effective Sample Size: 499
Number of Estimated Parameters: 4
Number of Constrained Parameters: 0
LogLikelihood: -717.972
AIC: 1443.945
BIC: 1460.795
Submodels
Estimate StandardError TStatistic PValue
________ _____________ __________ __________
State 1 Constant(1) 0.034171 0.067052 0.50963 0.61031
State 1 AR{1}(1,1) 0.14564 0.077043 1.8904 0.0587
State 2 Constant(1) 1.8688 0.22499 8.3063 9.8696e-17
State 2 AR{1}(1,1) 0.41827 0.064622 6.4726 9.6348e-11
Fit TAR Model with Exogenous Threshold Variable
The estimation of AR parameters in a self-exciting model is complicated by the mixed endogenous dynamics.
Compare SETAR model estimation to estimation of a TAR model, with the same general structure as the SETAR model, but with an exogenous threshold variable. Consider the threshold variable , where is standard Gaussian noise.
Simulate a random path of threshold variable observations, and then plot the threshold transition with the exogenous threshold data.
z = 1.5 + 5*randn(T,1); % Path of exogenous threshold data
figure
ttplot(ttDGP,Data=z)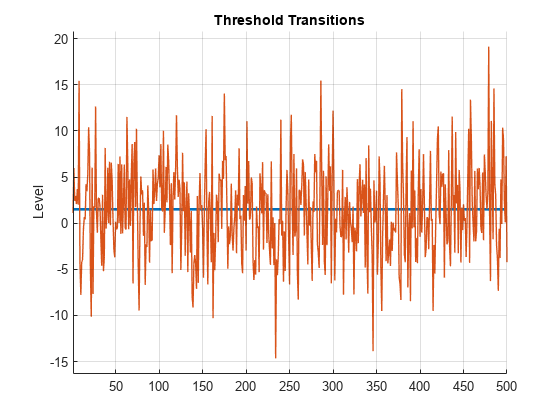
As expected, randomly fluctuates around the threshold.
Simulate a response path from the DGP of the TAR model. Specify that the threshold variable is exogenous and specify the simulated threshold data.
rng(seed) Y = simulate(MdlDGP,T,Type="exogenous",Z=z); % TAR path
Estimate the TAR model with an exogenous threshold variable. Threshold variable aside, the structure of the SETAR and TAR models are identical. Therefore, specify the SETAR model template and initial threshold transition. Specify that the threshold variable is exogenous and specify the simulated threshold data and the maximum transition rate maxRate.
EstMdlExo = estimate(Mdl,tt0,Y,MaxRate=maxRate, ... Type="exogenous",Z=z);
Visually compare the estimated parameters to those of the DGP.
EstMdlExoParams = [EstMdlExo.Switch.Levels; ... EstMdlExo.Submodels(1).Constant; ... EstMdlExo.Submodels(2).Constant; ... EstMdlExo.Submodels(1).AR{:}; ... EstMdlExo.Submodels(2).AR{:}; ... EstMdlExo.Covariance]; table(DGPParams,EstMdlParams,EstMdlExoParams)
ans=6×3 table
DGPParams EstMdlParams EstMdlExoParams
_________ ____________ _______________
1.5 1.5233 1.5391
0 0.034171 0.0048078
2 1.8688 1.9463
0.2 0.14564 0.18852
0.4 0.41827 0.41886
1 1.0406 1.0216
Overall, the estimates improve with the exogenous threshold data.
Summarize the estimation results.
summarize(EstMdlExo)
Description
1-Dimensional tsVAR Model with 2 Submodels
Switch
Transition Type: discrete
Estimated Levels: 1.539
Fit
Effective Sample Size: 499
Number of Estimated Parameters: 4
Number of Constrained Parameters: 0
LogLikelihood: -713.386
AIC: 1434.773
BIC: 1451.623
Submodels
Estimate StandardError TStatistic PValue
_________ _____________ __________ ___________
State 1 Constant(1) 0.0048078 0.08187 0.058726 0.95317
State 1 AR{1}(1,1) 0.18852 0.039812 4.7352 2.1887e-06
State 2 Constant(1) 1.9463 0.087457 22.254 1.0253e-109
State 2 AR{1}(1,1) 0.41886 0.040425 10.361 3.7112e-25
Tune Estimation Procedure
The MaxLevel and MaxRate name-value arguments set inequality constraints on the threshold parameter search. Estimate the TAR model with an exogenous threshold variable again. Expand the search domain by increasing the upper bound of the rate.
maxRate = 50;
EstMdlExo2 = estimate(Mdl,tt0,Y,MaxRate=maxRate,Type="exogenous",Z=z);Compare the results of the two MaxRate settings for the TAR model esimtation.
EstMdlExoParams2 = [EstMdlExo2.Switch.Levels; ... EstMdlExo2.Submodels(1).Constant; ... EstMdlExo2.Submodels(2).Constant; ... EstMdlExo2.Submodels(1).AR{:}; ... EstMdlExo2.Submodels(2).AR{:}; ... EstMdlExo2.Covariance]; table(DGPParams,EstMdlParams,EstMdlExoParams2)
ans=6×3 table
DGPParams EstMdlParams EstMdlExoParams2
_________ ____________ ________________
1.5 1.5233 1.5185
0 0.034171 0.0066017
2 1.8688 1.9409
0.2 0.14564 0.18838
0.4 0.41827 0.41627
1 1.0406 1.0063
In this case, the expanded search domain results in more accurate estimates.
Fit SESTAR Model Containing Regression Component
Accurate estimation is more challenging when the number of series, thresholds, and submodels increases for the following reasons:
The estimates are asymptotic.
When the estimation procedure estimates submodel parameters, the demand on the sample size increases because the procedure divides the sample into subsamples through transition-function weighting.
Consider a DGP for a bivariate, two-state, self-exciting, smooth TAR model (SESTAR) with both endogenous and exogenous dynamics:
The smooth, normal threshold transition is at mid-level 2.5 with a transition rate of 5.
The state 1 submodel is .
The state 2 submodel is .
and are univariate, standard Gaussian exogenous series.
and .
Create a SETAR model that represents the DGP by following this procedure. Use varm to specify the submodel dynamics.
ttDGP = threshold(2.5,Type="normal",Rates=5); % Constants (numSeries x 1 vectors) c1 = [1;-1]; c2 = [2;-2]; % AR coefficients (numSeries x numSeries matrices) AR1 = {[0.5 0; 0 0.5]}; % 1 lag AR2 = {[0.3 0; 0 0.3] [0.4 0; 0 0.4]}; % 2 lags % Regression coefficients (numSeries x numRegressors matrices) beta1 = [0.3; -0.3]; % 1 regressor Beta2 = [0.5 0.2; -0.5 -0.2]; % 2 regressors % Innovations covariances (numSeries x numSeries matrices) Sigma1 = [1 0; 0 1]; Sigma2 = [2 0; 0 2]; mdl1DGP = varm(Constant=c1,AR=AR1,Beta=beta1,Covariance=Sigma1); mdl2DGP = varm(Constant=c2,AR=AR2,Beta=Beta2,Covariance=Sigma2); MdlDGP = tsVAR(ttDGP,[mdl1DGP mdl2DGP]);
Simulate a 2-D path of standard Gaussian values for the regression data, and then simulate a 2-D response path from the DGP. By default, tsVAR object functions set the threshold variable of a multivariate self-exciting model to the first variable (Index=1).
rng(seed) X = rand(T,2); % Regression data Y = simulate(MdlDGP,T,X=X); % SESTAR path figure ttplot(ttDGP,Data=Y(:,1)) % Self-exciting component of Y colorbar('off')

The plot indicates that the self-exciting first component of the response samples states on either side of the threshold level.
Unconstrained Estimation
Estimate the two threshold parameters and all 22 constant, AR, and regression submodel parameters from the simulated sample. To show the progress of the search procedure, create an iteration plot by using the IterationPlot name-value argument.
tt = threshold(NaN,Type="normal",Rates=NaN); mdl1 = varm(Constant=NaN(2,1),AR={NaN(2)},Beta=NaN(2,1)); mdl2 = varm(Constant=NaN(2,1),AR={NaN(2) NaN(2)},Beta=NaN(2,2)); Mdl = tsVAR(tt,[mdl1 mdl2]); figure tt0 = threshold(1,Type="normal",Rates=1); EstMdlSESTAR = estimate(Mdl,tt0,Y,X=X,IterationPlot=true);

The plot indicates convergence after 20 iterations.
Compare the DGP parameters and estimated values.
compareEstimates(MdlDGP,EstMdlSESTAR)
Threshold parameters:
2.5000 2.6484
5.0000 4.6065
Submodel constants:
1.0000 1.4785
-1.0000 -1.2181
2.0000 1.0724
-2.0000 -1.7798
Submodel(1) AR coefficients:
0.5000 0 0.4550 0.0710
0 0.5000 0.0482 0.4903
Submodel(2) AR coefficients:
0.3000 0 0.3327 -0.1410
0 0.3000 -0.1514 0.2909
0.4000 0 0.3539 0.4015
0 0.4000 0.5119 0.3694
Submodel(1) regression coefficients:
0.3000 -0.0039
-0.3000 -0.0400
Submodel(2) regression coefficients:
0.5000 0.2000 0.3600 0.2931
-0.5000 -0.2000 -0.4952 0.2515
Innovations covariances:
1.0000 0 1.1646 0.0744
0 1.0000 0.0744 0.9572
2.0000 0 1.8074 0.1777
0 2.0000 0.1777 1.8293
In addition to the large number of parameters to estimate, the estimation procedure can struggle to distinguish the mix of self-exciting, AR, and exogenous dynamics. The results show a range of accuracies relative to the DGP, depending on the parameter.
Summarize the estimation results:
summarize(EstMdlSESTAR)
Description
2-Dimensional tsVAR Model with 2 Submodels
Switch
Transition Type: normal
Estimated Levels: 2.648
Estimated Rates: 4.607
Fit
Effective Sample Size: 498
Number of Estimated Parameters: 22
Number of Constrained Parameters: 0
LogLikelihood: -1587.781
AIC: 3219.561
BIC: 3312.195
Submodels
Estimate StandardError TStatistic PValue
__________ _____________ __________ __________
State 1 Constant(1) 1.4785 0.25733 5.7455 9.1639e-09
State 1 Constant(2) -1.2181 0.24873 -4.8972 9.7196e-07
State 1 AR{1}(1,1) 0.45497 0.098613 4.6137 3.9548e-06
State 1 AR{1}(2,1) 0.048204 0.095317 0.50573 0.61305
State 1 AR{1}(1,2) 0.071037 0.060538 1.1734 0.24062
State 1 AR{1}(2,2) 0.49026 0.058515 8.3784 5.363e-17
State 1 Beta(1,1) -0.0039272 0.27741 -0.014157 0.9887
State 1 Beta(2,1) -0.039972 0.26814 -0.14907 0.8815
State 2 Constant(1) 1.0724 0.40609 2.6408 0.0082704
State 2 Constant(2) -1.7798 0.39252 -4.5344 5.7775e-06
State 2 AR{1}(1,1) 0.33272 0.073856 4.505 6.6378e-06
State 2 AR{1}(2,1) -0.15145 0.071387 -2.1215 0.033883
State 2 AR{1}(1,2) -0.14097 0.054465 -2.5882 0.0096479
State 2 AR{1}(2,2) 0.29089 0.052645 5.5255 3.2851e-08
State 2 AR{2}(1,1) 0.35391 0.058696 6.0294 1.6452e-09
State 2 AR{2}(2,1) 0.51186 0.056735 9.022 1.8471e-19
State 2 AR{2}(1,2) 0.40149 0.054702 7.3395 2.1443e-13
State 2 AR{2}(2,2) 0.36939 0.052874 6.9863 2.8231e-12
State 2 Beta(1,1) 0.36 0.28804 1.2498 0.21136
State 2 Beta(2,1) -0.49523 0.27841 -1.7788 0.075276
State 2 Beta(1,2) 0.29314 0.2962 0.98967 0.32234
State 2 Beta(2,2) 0.25154 0.2863 0.87859 0.37962
Constrained Estimation
estimate can set equality constraints on theoretically or empirically known or hypothesized parameter values. Consider estimating the same SESTAR model using the same data, but constrain constants and regression coefficients to specific values.
tt = threshold(NaN,Type="normal",Rates=NaN); mdl1 = varm(Constant=[1;-1],AR={NaN(2)},Beta=[0.3; -0.3]); mdl2 = varm(Constant=[2;-2],AR={NaN(2) NaN(2)},Beta=[0.5 0.2; -0.5 -0.2]); Mdl = tsVAR(tt,[mdl1 mdl2]); figure tt0 = threshold(1,Type="normal",Rates=1); EstMdlCon = estimate(Mdl,tt0,Y,X=X,IterationPlot=true);
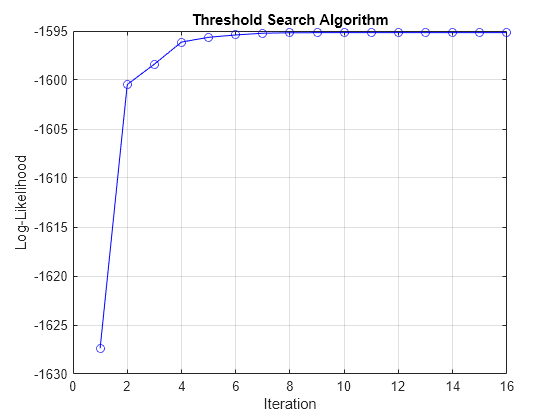
The plot indicates faster convergence, but at a reduced loglikelihood.
Compare the DGP and estimated threshold transitions.
varnames = ["DGP" "Unconstrained" "Constrained"]; array2table([MdlDGP.Switch.Levels, EstMdlSESTAR.Switch.Levels, EstMdlCon.Switch.Levels; ... MdlDGP.Switch.Rates, EstMdlSESTAR.Switch.Rates, EstMdlCon.Switch.Rates], ... VariableNames=varnames,RowNames=["MidLevel" "Rate"])
ans=2×3 table
DGP Unconstrained Constrained
___ _____________ ___________
MidLevel 2.5 2.6484 2.7977
Rate 5 4.6065 1.4487
The estimation results are mixed. For example, the threshold parameter search appears to be inhibited by the equality constraints, resulting in less accurate estimates.
Compare the DGP and estimated AR parameters.
ARSubmodel1 = table(MdlDGP.Submodels(1).AR{1},EstMdlSESTAR.Submodels(1).AR{1}, ...
EstMdlCon.Submodels(1).AR{1},VariableNames=varnames)ARSubmodel1=2×3 table
DGP Unconstrained Constrained
__________ ____________________ _____________________
0.5 0 0.45497 0.071037 0.47106 -0.012461
0 0.5 0.048204 0.49026 0.066586 0.51035
ARSubmodel2 = table([MdlDGP.Submodels(2).AR{1}; MdlDGP.Submodels(2).AR{2}], ...
[EstMdlSESTAR.Submodels(2).AR{1}; EstMdlSESTAR.Submodels(2).AR{2}], ...
[EstMdlCon.Submodels(2).AR{1}; EstMdlCon.Submodels(2).AR{2}], ...
VariableNames=varnames)ARSubmodel2=4×3 table
DGP Unconstrained Constrained
__________ ____________________ ____________________
0.3 0 0.33272 -0.14097 0.18716 -0.10112
0 0.3 -0.15145 0.29089 -0.12179 0.25292
0.4 0 0.35391 0.40149 0.33386 0.45529
0 0.4 0.51186 0.36939 0.56162 0.37272
The AR parameter estimates show a mix of both improved and reduced accuracy under the equality constraints.
Summarize the estimation results.
summarize(EstMdlCon)
Description
2-Dimensional tsVAR Model with 2 Submodels
Switch
Transition Type: normal
Estimated Levels: 2.798
Estimated Rates: 1.449
Fit
Effective Sample Size: 498
Number of Estimated Parameters: 12
Number of Constrained Parameters: 10
LogLikelihood: -1595.169
AIC: 3214.338
BIC: 3264.865
Submodels
Estimate StandardError TStatistic PValue
_________ _____________ __________ __________
State 1 Constant(1) 1 0 Inf 0
State 1 Constant(2) -1 0 -Inf 0
State 1 AR{1}(1,1) 0.47106 0.075253 6.2597 3.8577e-10
State 1 AR{1}(2,1) 0.066586 0.072189 0.92239 0.35633
State 1 AR{1}(1,2) -0.012461 0.042098 -0.29599 0.76724
State 1 AR{1}(2,2) 0.51035 0.040384 12.638 1.3114e-36
State 1 Beta(1,1) 0.3 0 Inf 0
State 1 Beta(2,1) -0.3 0 -Inf 0
State 2 Constant(1) 2 0 Inf 0
State 2 Constant(2) -2 0 -Inf 0
State 2 AR{1}(1,1) 0.18716 0.057629 3.2476 0.0011638
State 2 AR{1}(2,1) -0.12179 0.055283 -2.203 0.027594
State 2 AR{1}(1,2) -0.10112 0.05479 -1.8455 0.06496
State 2 AR{1}(2,2) 0.25292 0.05256 4.8121 1.4932e-06
State 2 AR{2}(1,1) 0.33386 0.060951 5.4776 4.3122e-08
State 2 AR{2}(2,1) 0.56162 0.058469 9.6054 7.5852e-22
State 2 AR{2}(1,2) 0.45529 0.057332 7.9413 2.001e-15
State 2 AR{2}(2,2) 0.37272 0.054997 6.7771 1.2259e-11
State 2 Beta(1,1) 0.5 0 Inf 0
State 2 Beta(2,1) -0.5 0 -Inf 0
State 2 Beta(1,2) 0.2 0 Inf 0
State 2 Beta(2,2) -0.2 0 -Inf 0
Likelihood surfaces for the mixture densities of switching models, such as the negative sum-of-squares function evaluated by estimate, can have steep ridges and flat basins with highly contrasting directional derivatives, leading search algorithms to local maxima [2] [3]. In general, estimate is sensitive to the following characteristics:
How well the model describes the data
How well the data samples the submodels
How large the sample size is
How many parameters are estimated
How the search is initiated and constrained
In practice, you can control only some of these parameters, and heuristic adjustments that lead to improved estimates are case-dependent.
Supporting Functions
This function displays all SESTAR model DGP values and estimates.
function compareEstimates(MdlDGP,EstMdl) fprintf("\nThreshold parameters:\n") disp([MdlDGP.Switch.Levels EstMdl.Switch.Levels; ... MdlDGP.Switch.Rates EstMdl.Switch.Rates]) fprintf("\n\nSubmodel constants:\n") disp([MdlDGP.Submodels(1).Constant EstMdl.Submodels(1).Constant; ... MdlDGP.Submodels(2).Constant EstMdl.Submodels(2).Constant]) fprintf("\n\nSubmodel(1) AR coefficients:\n") disp([MdlDGP.Submodels(1).AR{:} EstMdl.Submodels(1).AR{:}]) fprintf("\n\nSubmodel(2) AR coefficients:\n") disp([MdlDGP.Submodels(2).AR{1} EstMdl.Submodels(2).AR{1}; ... MdlDGP.Submodels(2).AR{2} EstMdl.Submodels(2).AR{2}]) fprintf("\n\nSubmodel(1) regression coefficients:\n") disp([MdlDGP.Submodels(1).Beta EstMdl.Submodels(1).Beta]) fprintf("\n\nSubmodel(2) regression coefficients:\n") disp([MdlDGP.Submodels(2).Beta EstMdl.Submodels(2).Beta]) fprintf("\n\nInnovations covariances:\n") disp([MdlDGP.Submodels(1).Covariance EstMdl.Submodels(1).Covariance; ... MdlDGP.Submodels(2).Covariance EstMdl.Submodels(2).Covariance]) end
References
[2] Hamilton, James D. Time Series Analysis. Princeton, NJ: Princeton University Press, 1994.