This example requires an internet connection.
Open a new instance of the Genomics Viewer app. Specify to use Human (GRCh38/hg38) as the reference sequence and the corresponding cytoband ideogram file.
Alternatively, you can use the setReference function to specify the reference sequence and the cytoband file.
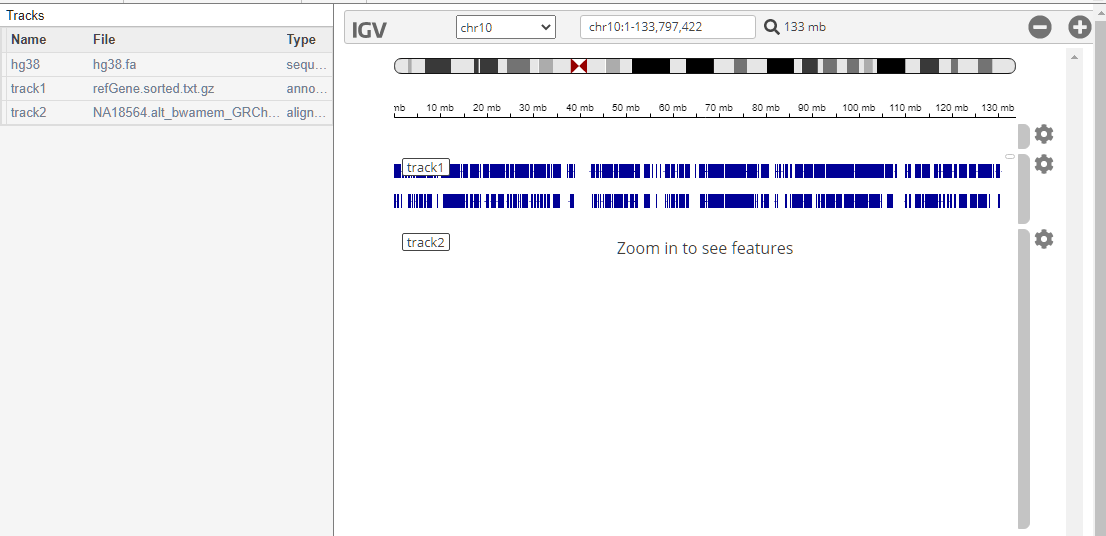
In the IGV panel, click Chromosome 10 to zoom in on it.
The IGV panel is updated as follows.
Add the Refseq genes as the annotation file.
Load a low coverage alignment data. The sample ID is NA18564 and the sample has been identified with the CYP2C19*3 mutation.
To center the alignment data around the location of the mutation on the CYP2C19 gene, enter chr10:94,780,635-94,780,673 in the search text box.

To show the bases on track2, click the gear icon and select Show all bases.
The IGV panel is updated as follows.
For additional information on other interactive visualization options, see Visualize NGS Data Using Genomics Viewer App.
Close the app.