Register Multimodal Medical Image Volumes with Spatial Referencing
This example shows how to align two medical image volumes using moment-of-mass-based registration.
Multimodal image registration aligns images acquired using different imaging modalities, such as MRI and CT. Even when acquired for the same patient and region of interest, multimodal images can be misaligned due to differences in patient positioning (translation or rotation) and voxel size (scaling). Different imaging modalities often have different voxel sizes due to scanner variability and concerns about scan time and radiation dose. The imregmoment function enables fast registration of 3-D image volumes, including multimodal images.
Medical images have an intrinsic coordinate system defined by rows, columns, and slices and a patient coordinate system that describes the real-world position and voxel spacing. You can use the medicalVolume object to import and manage the spatial referencing of a medical image volume. To register images with different voxel sizes using imregmoment, you must use spatial referencing to register the images in the patient coordinate system. During registration, imregmoment aligns a moving image with a fixed image, and then resamples the aligned image in the intrinsic coordinates of the fixed image. The registered volumes share one patient coordinate system and have the same number of rows, columns, and slices.
In this example, you use medicalVolume and imregmoment to register multimodal MRI and CT images of the head.
Load Images
The data used in this example is a modified version of the 3-D CT and MRI data sets from The Retrospective Image Registration Evaluation (RIRE) Dataset, provided by Dr. Michael Fitzpatrick. For more information, see the RIRE Project homepage. The modified data set contains one CT scan and one MRI scan stored in the NRRD file format. The size of the entire data set is approximately 35 MB. Download the data set from the MathWorks® website, then unzip the folder.
zipFile = matlab.internal.examples.downloadSupportFile("medical", ... "MedicalRegistrationNRRDdata.zip"); filepath = fileparts(zipFile); unzip(zipFile,filepath)
In image registration, consider one image to be the fixed image and the other image to be the moving image. The goal of registration is to align the moving image with the fixed image. In this example, the fixed image is a T1 weighted MRI image. The moving image is a CT image from the same patient. The images are stored in the NRRD file format.
Use medicalVolume to read the MRI image. By looking at the Voxels and VoxelSpacing properties, you can determine that the MRI volume is a 256-by-256-by-26 voxel array, where each voxel is 1.25-by-1.25-by-4.0 mm.
filenameMRI = fullfile(filepath,"supportfilesNRRD","Patient007MRT1.nrrd"); fixedMRIVolume = medicalVolume(filenameMRI)
fixedMRIVolume =
medicalVolume with properties:
Voxels: [256×256×26 single]
VolumeGeometry: [1×1 medicalref3d]
SpatialUnits: "unknown"
Orientation: "unknown"
VoxelSpacing: [1.2500 1.2500 4]
NormalVector: [0 0 1]
NumCoronalSlices: []
NumSagittalSlices: []
NumTransverseSlices: []
PlaneMapping: ["unknown" "unknown" "unknown"]
Modality: "unknown"
WindowCenters: []
WindowWidths: []
Import the CT image. The size of each voxel in the CT image is 0.65-by-0.65-by-4 mm, which is smaller than in the MRI image.
filenameCT = fullfile(filepath,"supportfilesNRRD","Patient007CT.nrrd"); movingCTVolume = medicalVolume(filenameCT)
movingCTVolume =
medicalVolume with properties:
Voxels: [512×512×28 single]
VolumeGeometry: [1×1 medicalref3d]
SpatialUnits: "unknown"
Orientation: "unknown"
VoxelSpacing: [0.6536 0.6536 4]
NormalVector: [0 0 1]
NumCoronalSlices: []
NumSagittalSlices: []
NumTransverseSlices: []
PlaneMapping: ["unknown" "unknown" "unknown"]
Modality: "unknown"
WindowCenters: []
WindowWidths: []
Extract the image voxel data from the Voxels property of the medicalVolume objects.
fixedMRIVoxels = fixedMRIVolume.Voxels; movingCTVoxels = movingCTVolume.Voxels;
Display Unregistered Images
Use the imshowpair function to judge image alignment. First, calculate the location of the middle slice of each volume. The VolumeGeometry property of the medical volume object contains a medicalref3d object, which specifies spatial details about the volume. Use the VolumeSize property of each medicalref3d object to calculate the location of the middle slice of the corresponding volume, in voxels.
fixedVolumeSize = fixedMRIVolume.VolumeGeometry.VolumeSize; movingVolumeSize = movingCTVolume.VolumeGeometry.VolumeSize; centerFixed = fixedVolumeSize/2; centerMoving = movingVolumeSize/2;
Define spatial referencing for the 2-D slices using imref2d. Specify the size, in voxels, and voxel spacing, in millimeters, of the transverse slices.
fixedVoxelSpacing = fixedMRIVolume.VoxelSpacing; movingVoxelSpacing = movingCTVolume.VoxelSpacing; Rfixed2D = imref2d(fixedVolumeSize(1:2),fixedVoxelSpacing(2),fixedVoxelSpacing(1)); Rmoving2D = imref2d(movingVolumeSize(1:2),movingVoxelSpacing(2),movingVoxelSpacing(1));
Display the image slices in the patient coordinate system using imshowpair. Plot the slice in the third spatial dimension, which corresponds to the transverse anatomical plane. The MRI slice is magenta, and the CT slice is green.
figure imshowpair(movingCTVoxels(:,:,centerMoving(3)), ... Rmoving2D, ... fixedMRIVoxels(:,:,centerFixed(3)), ... Rfixed2D) title("Unregistered Transverse Slice")

You can also display the volumes as 3-D objects. Create a viewer3d object, in which you can display multiple volumes.
viewerUnregistered = viewer3d(BackgroundColor="black",BackgroundGradient="off");
Display the medicalVolume objects as 3-D isosurfaces using volshow. The volshow function uses medicalVolume properties to display each volume in its respective patient coordinate system.
volshow(fixedMRIVolume, ... Parent=viewerUnregistered, ... RenderingStyle="Isosurface", ... IsosurfaceValue=0.05, ... Colormap=[1 0 1], ... Alphamap=1);
Warning: The volume's VolumeGeometry.PatientCoordinateSystem property is not set. Assuming "LPS+" for visualization.
volshow(movingCTVolume, ... Parent=viewerUnregistered, ... RenderingStyle="Isosurface", ... IsosurfaceValue=0.05, ... Colormap=[0 1 0], ... Alphamap=1);
Warning: The volume's VolumeGeometry.PatientCoordinateSystem property is not set. Assuming "LPS+" for visualization.

Register Images
To accurately register volumes with different voxel dimensions, specify the spatial referencing for each volume using imref3d objects. Create each imref3d object using the volume size in voxels and the voxel dimensions in millimeters.
Rfixed3d = imref3d(fixedVolumeSize,fixedVoxelSpacing(2), ... fixedVoxelSpacing(1),fixedVoxelSpacing(3)); Rmoving3d = imref3d(movingVolumeSize,movingVoxelSpacing(2), ... movingVoxelSpacing(1),movingVoxelSpacing(3));
The imregmoment function sets fill pixels added to the registered volume to 0. To improve the display of registration results, scale the CT intensities to the range [0, 1], so that the fill value is equal to the minimum of the image data range.
rescaledMovingCTVoxels = rescale(movingCTVoxels);
Register the volumes using imregmoment, including the imref3d objects as inputs. Specify the MedianThresholdBitmap name-value argument as true, which is appropriate for multimodal images.
[geomtform,movingCTRegisteredVoxels] = imregmoment(rescaledMovingCTVoxels,Rmoving3d, ...
fixedMRIVoxels,Rfixed3d,MedianThresholdBitmap=true);The geomtform output is an affinetform3d geometric transformation object. The T property of geomtform contains the 3-D affine transformation matrix that maps the moving CT volume in its patient coordinate system to the fixed MRI volume in its patient coordinate system.
geomtform.T
ans = 4×4
1.0000 -0.0039 0.0013 0
0.0039 1.0000 0.0063 0
-0.0014 -0.0063 1.0000 0
-4.8094 -16.0063 -1.3481 1.0000
The movingRegisteredVoxels output contains the registered CT volume. The imregmoment function resamples the aligned CT volume in the MRI intrinsic coordinates, so the registered volumes have the same number of rows, columns, and slices.
whos movingCTRegisteredVoxels fixedMRIVoxels
Name Size Bytes Class Attributes fixedMRIVoxels 256x256x26 6815744 single movingCTRegisteredVoxels 256x256x26 6815744 single
Create medicalVolume Object for Registered Image
Create a new medicalVolume object that contains the registered voxel data and its spatial referencing information. You can create a medicalVolume object by specifying a voxel array and a medicalref3d object. The registered CT volume has the same spatial referencing as the original MRI volume, so use the medicalref3d object stored in the VolumeGeometry property of fixedMRIVolume.
R = fixedMRIVolume.VolumeGeometry; movingRegisteredVolume = medicalVolume(movingCTRegisteredVoxels,R)
movingRegisteredVolume =
medicalVolume with properties:
Voxels: [256×256×26 single]
VolumeGeometry: [1×1 medicalref3d]
SpatialUnits: "unknown"
Orientation: "unknown"
VoxelSpacing: [1.2500 1.2500 4]
NormalVector: [0 0 1]
NumCoronalSlices: []
NumSagittalSlices: []
NumTransverseSlices: []
PlaneMapping: ["unknown" "unknown" "unknown"]
Modality: "unknown"
WindowCenters: []
WindowWidths: []
Display Registered Images
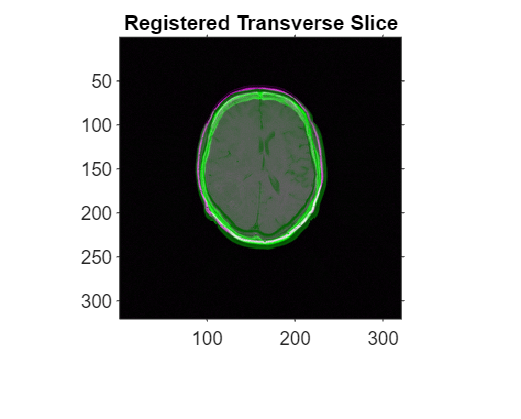
To check the registration results, use imshowpair to view the middle transverse slices of the fixed and registered volumes. Use the spatial referencing object for the fixed volume.
figure imshowpair(movingRegisteredVolume.Voxels(:,:,centerFixed(3)), ... Rfixed2D,fixedMRIVoxels(:,:,centerFixed(3)),Rfixed2D) title("Registered Transverse Slice")
Display the registered volumes as 3-D isosurfaces using volshow. You can zoom and rotate the display to assess the registration results.
viewerRegistered = viewer3d(BackgroundColor="black",BackgroundGradient="off"); volshow(fixedMRIVolume, ... Parent=viewerRegistered, ... RenderingStyle="Isosurface", ... IsosurfaceValue=0.05, ... Colormap=[1 0 1], ... Alphamap=1);
Warning: The volume's VolumeGeometry.PatientCoordinateSystem property is not set. Assuming "LPS+" for visualization.
volshow(movingRegisteredVolume, ... Parent=viewerRegistered, ... RenderingStyle="Isosurface", ... IsosurfaceValue=0.05, ... Colormap=[0 1 0], ... Alphamap=1);
Warning: The volume's VolumeGeometry.PatientCoordinateSystem property is not set. Assuming "LPS+" for visualization.

See Also
imregmoment | medicalref3d | medicalVolume